Abstract
Identifying genes that regulate bone marrow (BM) engraftment may reveal molecular targets for overcoming engraftment barriers. To achieve this aim, we applied a forward genetic approach in a mouse model of nonmyeloablative BM transplantation. We evaluated engraftment of allogeneic and syngeneic BM in BALB.K and B10.BR recipients. This allowed us to partition engraftment resistance into its intermediate phenotypes, which are firstly the immune-mediated resistance and secondly the nonimmune rejection of donor BM cells. We observed that BALB.K and B10.BR mice differed with regard to each of these resistance mechanisms, thereby providing evidence that both are under genetic control. We then generated a segregating backcross (n = 200) between the BALB.K and B10.BR strains to analyze for genetic linkage to the allogeneic BM engraftment phenotype using a 127-marker genome scan. This analysis identified a novel quantitative trait locus (QTL) on chromosome 16, termed Bmgr5 (logarithm of odds 6.4, at 11.1 cM). The QTL encodes susceptibility alleles, from the BALB.K strain, that are permissive for allogeneic BM engraftment. Further identification of Bmgr5 genes by positional cloning may reveal new and effective approaches for overcoming BM engraftment obstacles.
Introduction
Understanding engraftment barriers is central to successful allogeneic hematopoietic cell transplantation. These barriers are difficult to study because multiple cell types, molecular pathways, and mechanisms act in concert. Both host natural killer (NK) cells and T cells, for example, can mediate resistance to engraftment.1,2 Elimination of donor hematopoietic cells by these immune cell populations likely requires cytolytic mechanisms, such as granzyme and perforin. However, key cytolytic pathways have not been identified, because neutralization of each singly,3,4 or even several simultaneously,5,6 fails to eliminate engraftment barriers. In addition, nonimmune elements related to bone marrow (BM) “space” and hematopoietic stem cell (HSC) niche interactions likely contribute to engraftment barriers7,8 and further confound efforts to study their underlying biology.
Genetic studies of disease and physiologic traits may provide key insights into molecular mechanisms and lead to novel therapy.9,10 With regard to hematopoietic engraftment barriers, it is established that polymorphisms of genes encoding histocompatibility antigens are causal in activating cellular responses that mediate graft rejection.11 Other than this understanding, however, little is known about the genetic regulation of engraftment resistance. We addressed this problem by applying a forward genetic approach using a new mouse model of nonmyeloablative BM transplantation. We first characterized donor chimerism, tolerance, and immune resistance mechanisms to show that our model shares all features of nonmyeloablative allogeneic BM engraftment in rodents. We next compared allogeneic and syngeneic BM engraftment to further model, respectively, immune-mediated resistance and nonimmune rejection of hematopoietic cells. From a genetic perspective, these resistance mechanisms can be said to represent the 2 intermediate phenotypes that constitute the overall BM rejection trait. We provide evidence that both are under genetic control by demonstrating strain-specific variation, between BALB.K and B10.BR mice, in these engraftment attributes. We then used a segregating backcross (BC) generated from these 2 strains for genetic linkage analysis and identified a novel quantitative trait locus (QTL) on proximal chromosome 16, termed Bmgr5. This QTL encodes susceptibility genes that confer permissiveness for allogeneic BM engraftment in these mice. These results provide the groundwork for positional cloning of BM engraftment resistance genes as a prerequisite for novel approaches aimed at overcoming engraftment barriers for hematopoietic cell transplantation.
Methods
Animal facilities and equipment
This study was initiated in the laboratory of J.A.S. and the Research Animal Facility at Stanford University, where the BM engraftment phenotype was characterized. The study was completed in the laboratory of T.M.C. and the Animal Resource Center at the University of Utah, where genetic characterization of the model in F1 and BC hybrid mice occurred. Total body irradiation (TBI) was delivered using a Philips Unit Irradiator) at Stanford or an X-RAD 320 Biological Irradiator (Precision X-Ray) at Utah. All work performed using animals was reviewed and approved by the Administrative Panel on Laboratory Animal Care at Stanford University and the Institutional Animal Care and Use Committee at the University of Utah.
Mouse strains and crosses
AKR/J (H2k, Thy1.1) mice served as BM and HSC donors. BALB.K (H2k, Thy1.2) and B10.BR (H2k, Thy1.2) mice were used as transplant recipients. C3H/Hen (H2k) mice served as third-party donors in cardiac allograft experiments. BALB.K female and B10.BR male mice were mated to generate a (B10.BR × BALB.K)F1 population. F1 mice were subsequently mated with females of the B10.BR parental strain to produce [B10.BR × (B10.BR × BALB.K)F1] BC mice for linkage analysis.
BM and HSC isolation
BM was harvested from donor mice after CO2 asphyxiation. In some experiments, a population enriched for long-term HSCs were isolated from BM as previously described.12 In brief, c-Kit (3C11) positive BM cells were selected via micromagnetic bead separation for multiparameter FACS sorting of a cKit+Thy1lo−intLin−Sca1+ composite immunophenotype.
BM and HSC transplantation
BALB.K and B10.BR recipient mice were conditioned with nonmyeloablative TBI delivered in a single fraction on day 0. For some experiments, recipient mice were also injected intraperitoneally with 1500 μg anti-CD4 (GK1.5) and/or 1500 μg anti-CD8 (53-6.7) monoclonal antibodies given in 3 divided doses on days −3, −2, and −1; polyclonal anti–asialo-GM1 (Wako Chemical) 100 μg intravenously on day −7 and 100 μg intraperitoneally on day −1 as described3; or control rat IgG (Sigma-Aldrich). Anti-CD4 and anti-CD8 monoclonal antibodies were obtained by culturing the respective hybridomas in 15% fetal bovine serum in a CellLine flask-based bioreactor (Integra Biosciences) and purified by protein G (Amersham Biosciences) affinity chromatography.
Allogeneic chimerism analysis
Engraftment was evaluated in recipients of allogeneic BM or HSCs beginning at 6 weeks after transplantation. T-cell chimerism was assessed by FACS analysis using monoclonal antibodies against Thy1.1 (Ox-7) expressed by donor mice and Thy1.2 (53-2.1) expressed by recipient mice.
B-cell and myeloid chimerism was assessed by 1 of 2 techniques. In earlier studies, FACS sorted B220+ and Mac1/Gr1+ peripheral blood cells of transplanted mice were used for DNA extraction. As described, donor-derived cells were detected by genotyping for informative microsatellite markers.12 D6Mit3 and D8Mit224 were used for this purpose. Primers, polymorphisms, and PCR conditions are summarized in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
In later studies, B-cell and myeloid chimerism was measured by quantifying the relative expression of donor alleles in single nucleotide polymorphism (SNP) markers found on lineage-specific gene transcripts. A reverse transcription-polymerase chain reaction (RT-PCR) based method, adapted from procedures previously described,13 was used for this purpose. We identified and designed RT-PCR assays for 2 coding SNPs, rs47932153 and rs31178388, present, respectively, on Cd22 and Ncf2 mRNA (Table S1). Transcription of Cd22 and Ncf2 is highly enriched in murine B cells and myeloid cells, respectively, as we demonstrated by RT-PCR (data not shown). In brief, total RNA was extracted from spleen cells of transplanted mice using the RNeasy Mini Kit (QIAGEN) and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) for asymmetric PCR and high-resolution melting14 on a LightCycler 480 Instrument (Roche Applied Sciences). Donor chimerism was quantified, against a reference standard generated from control RNA as the ratio of donor to recipient melt curve peak height on the negative first-derivative plot of fluorescence with respect to temperature (dF/dT), as described in detail elsewhere.13
Syngeneic chimerism analysis
Engraftment in female mice given syngeneic male BM was evaluated by real-time PCR to quantify male DNA. PCR assays are summarized in supplemental Table 1. Syngeneic donor engraftment was quantified as the fold change in Sry expression, relative to control male DNA, by the comparative cycle threshold method using β-actin as an internal control.15
T-cell receptor Vβ analysis
FACS analysis of T-cell receptor Vβ subsets in spleen was performed at 2 and 12 months after transplantation in selected mice. Cells were labeled with anti-Vβ3 (KJ25), anti-Vβ4 (KT4), anti-Vβ5.1/5.2 (MR9-4), or anti-Vβ6 (RR4-7) and counterstained with either anti-Thy1.1 or anti-Thy1.2 for FACS analysis.
Intrapinna neonatal cardiac transplantation
Twelve to 16 weeks after transplantation, newborn heart grafts were placed into a pouch in the ear pinna of BM chimeric mice, according to the methods of Judd and Trentin as described.16 Heart grafts were visually monitored for contractions and survival was based on the time interval until contractions were no longer detectable.
Apoptosis and cell viability analysis
Apoptosis was determined by staining ethanol-fixed cells with propidium iodide for FACS analysis as described.17 The sub-G0/1 population was gated and used as the apoptotic population. Cell viability was assessed by trypan blue exclusion and enumerated using a hemocytometer cell counting chamber.
Genotyping
Genomic DNA was isolated from tail tips of BC mice using the DNeasy Tissue Kit (Qiagen). SNP genotyping was performed by PCR and high-resolution melt curve analysis on a LightCycler 480 Instrument (Roche Applied Sciences).14 A list of SNP markers used for genome scanning is provided as supplemental Table 2. Marker map position and order assignment were derived from linear interpolation against the Wellcome Trust mouse SNP genetic map.18 PCR assays are summarized in supplemental Table 3. BC mice were genotyped at each SNP marker as homozygous for B10.BR or heterozygous for B10.BR/BALB.K alleles according to the melt curve profile on the normalized and temperature-shifted fluorescence over temperature plot as previously described.14
Statistical and linkage analysis
A 127-marker genome scan was completed by genotyping 200 BC mice evaluated for the allogeneic BM engraftment trait. Ninety-one genotypes failed, yielding a genotype success rate of 99.7%. Linkage analysis for QTL mapping was performed using R/qtl version 1.10-27.19 The BM engraftment phenotype was analyzed as both a quantitative and binary trait. For evaluation as a quantitative trait, the percentage of donor T cells 8 weeks after transplantation was used as a continuous trait value. For analysis as a binary trait, mice were scored as having engrafted if any level of donor T cells was detected and engraftment in B-cell and myeloid lineages was demonstrated. Simple interval mapping was performed using the EM algorithm to test for maximum likelihood of single QTL effects calculated at 1-cM intervals along the genome.20 Marker imputation was not used to account for missing genotype data. Genome-wide significance thresholds were determined by empirical permutation testing.21 Standard genome-wide P values to define significant (P < .05) and highly significant (P < .001) threshold levels of logarithm of odds (LOD) scores for linkage were applied.22 For our BC mice, this corresponded to LOD scores of 2.7 and 4.3, respectively, based on results from 10 000 permutations. Approximate QTL confidence intervals were obtained using the 1.0-LOD drop-off method.23 Odds ratios (ORs) were derived from simple logistic regression using dichotomized engraftment values as the dependent variable and SNP marker genotype at linked QTL as categorical covariates.
Results
Characterization of the BM engraftment model
We initiated our studies by developing a new model of nonmyeloablative allogeneic BM transplantation. We used a donor mouse strain, AKR/J, and a major histocompatibility complex (MHC)–congenic recipient, BALB.K, that both share the H2k MHC haplotype. This is thus a MHC-identical and minor histocompatibility antigen-mismatched mouse strain combination that models clinically relevant unrelated and sibling donor allogeneic hematopoietic cell transplantation in humans. Rejection of long-term HSCs, short-term repopulating cells, and BM replete with nonprogenitor accessory cells may occur via distinct mechanisms. We therefore initially evaluated engraftment of a cKit+Thy1lo-intLin−Sca1+ stem and progenitor cell population alongside BM. Methods for detecting hematopoietic engraftment include spleen cell proliferation24 or progenitor cell assays,4 both of which measure early rejection. We chose to use the more stringent assay of long-term multilineage donor chimerism in peripheral compartments to evaluate for engraftment. Engraftment was defined by the presence of (1) donor T cells measured at 2 separate time points, beginning at 6 weeks after transplantation, and (2) donor-derived B-cells and myeloid cells at least once before the end of each experiment.
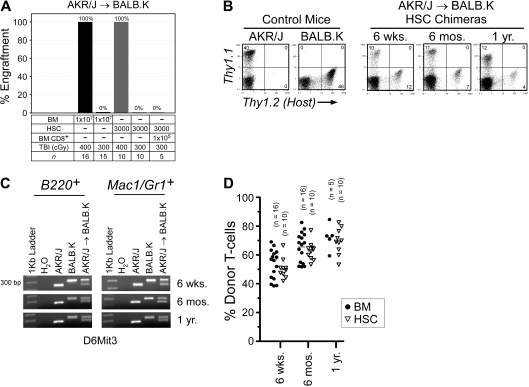
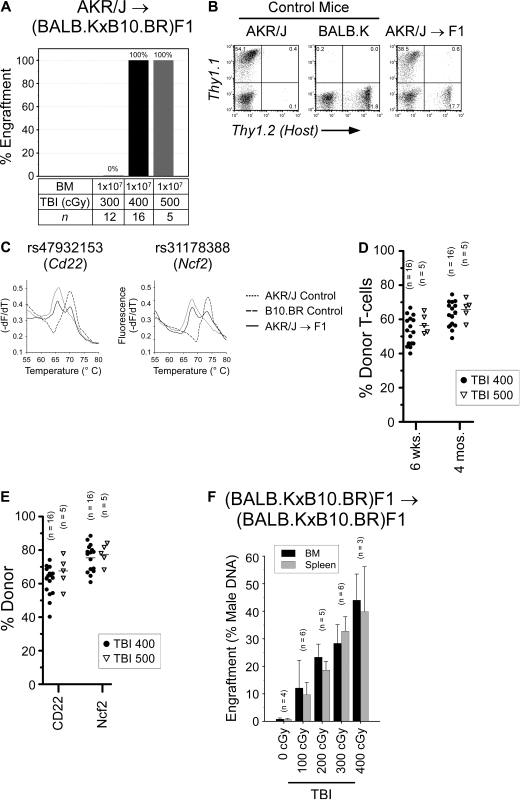
In AKR/J → BALB.K mice, we determined that TBI 400 cGy was the minimum dose that permitted BM engraftment (Figure 1A). We also transplanted HSCs and found a similar engraftment barrier. TBI 400 cGy is nonmyeloablative, as irradiated mice uniformly survived and had reconstituted autologous hematopoiesis in all blood lineages (data not shown). At the TBI 300 cGy dose that did not permit BM engraftment, we correspondingly found that coinjection of donor BM CD8α+ cells did not facilitate HSC engraftment. CD8α+ cells are the major HSC engraftment facilitating cell populations resident in BM in this strain combination25 as well as in MHC-mismatched mouse pairs.26 Engraftment was defined as the detection of donor cells in T-cell, B-cell, and myeloid cell compartments. We evaluated T-cell chimerism by FACS analysis of peripheral blood using the pan T-cell Thy1 marker (Figure 1B). B-cell and myeloid chimerism was evaluated by PCR of DNA, purified from FACS-sorted B220+ and Mac1/Gr1+ peripheral blood of transplanted mice, for a microsatellite marker polymorphic between donors and recipients (Figure 1C). As shown, donor engraftment was characterized by mixed trilineage chimerism and was durable to 1 year after transplantation (Figure 1D).
Figure 1.
Allogeneic BM engraftment in AKR/J → BALB.K (H2k) mice. Recipient BALB.K mice were treated with TBI 300 or 400 cGy and injected with BM or HSCs from MHC-matched AKR/J donors. (A) Percentage of mice with donor engraftment 6 weeks after transplantation. For engrafted mice, blood chimerism at indicated time points are shown with (B) representative FACS analysis for T cells using Thy1.1 (donor) and Thy1.2 (host) cell-surface markers, (C) representative PCR for B-cell and myeloid chimerism using DNA extracted from FACS-sorted B220+ and Mac1/Gr1+ blood cells amplified for the DNA marker D6Mit3, and (D) dot plot summarizing T-cell chimerism.
We next characterized the tolerant state induced after hematopoietic cell engraftment by analyzing the T-cell receptor Vβ repertoire. Vβ deletion mediated by immune responses to superantigens encoded by endogenous mouse mammary tumor virus genes allows inferences to be made regarding deletion of donor- versus host-derived T cells.27 Whereas only control AKR/J mice delete Vβ6, and BALB.K mice delete Vβ3, both delete Vβ5 (supplemental Figure 1). At 2 months after transplantation in AKR/J → BALB.K mixed chimeras, both donor- and host-derived Vβ4 splenic T cells had reconstituted to pretransplantation levels. From that time point extending until 12 months after transplantation, donor-derived Vβ3 T cells are essentially absent, as are host-derived Vβ6 T cells, indicating simultaneous deletion mediated by engrafted donor AKR/J HSCs as well as residual host BALB.K HSCs. These results suggest that central deletional tolerance, as reflected by deletion of both donor host-reactive and host donor-reactive T cells, contribute to the tolerant state in our model.
We then assayed for donor-specific organ tolerance in AKR/J → BALB.K mixed chimeras using neonatal cardiac grafts. In this assay cardiac allografts were harvested from neonates and heterotopically transplanted onto the external auditory pinna of recipient mice.16 AKR/J → BALB.K mixed chimeras were given neonatal cardiac grafts between 16 and 18 weeks after transplantation (Table 1). Third-party heart grafts from C3H/HeN donors were promptly rejected between 18 and 28 days after cardiac grafting. In contrast, both donor and control host-type heart grafts survived indefinitely until the end of the experiment. These results show that engraftment in our model induces specific systemic tolerance, allowing acceptance of donor solid organ allografts, in addition to permitting recovery of immune competence as reflected by rejection of third-party grafts.
Table 1.
Neonatal heart graft survival in AKR/J → BALB.K (H2) chimeric mice
| Heart graft | Graft survival, days* |
|---|---|
| AKR/J (n = 15) | > 100 × 15 |
| BALB.K (n = 7) | > 100 × 7 |
| C3H/HeN (n = 8) | 18 × 3, 25 × 2, 28 × 3 |
Neonatal heart grafts implanted 16 to 18 weeks after transplantation. Results pooled from 2 independent experiments.
Data are given as days × number of grafts.
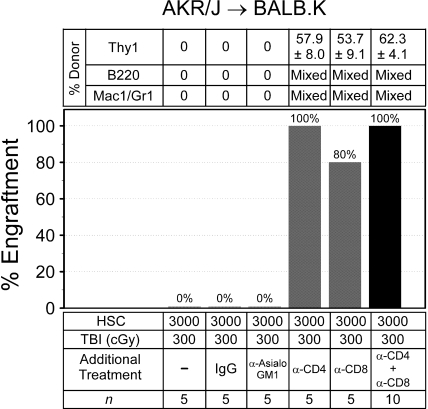
We evaluated the immune barrier to engraftment by adding depleting antibodies to TBI 300 cGy, the dose that did not permit engraftment, for recipient conditioning and transplanted donor HSCs (Figure 2). Administration of irrelevant rat IgG or the NK cell depleting anti-asialoGM1 reagent to TBI 300 cGy was not permissive for donor engraftment. The addition of depleting doses of anti-CD4 alone to TBI 300 cGy allowed engraftment in all mice, and 80% of the mice engrafted when anti-CD8 preceded TBI 300 cGy. Reliable engraftment was also achieved when anti-CD4 was combined with anti-CD8 before TBI 300 cGy. These data show that immune resistance in our mice—like other MHC-matched, minor histocompatibility antigen–mismatched mouse models28—is mediated primarily by host T cells.
Figure 2.
Immune barriers in AKR/J → BALB.K mice. Recipient mice were given sublethal TBI and injected with anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-asialoGM1, or control IgG before infusion of donor HSCs. Shown are the percentages of mice with donor engraftment and chimerism characteristics at 6 weeks after transplantation. T-cell chimerism was evaluated by FACS for Thy1 surface markers. B-cell and myeloid chimerism was evaluated by PCR for D6Mit3 DNA markers of FACS-sorted B220+ and Mac1/Gr1+ blood cells.
Collectively, these results show that our model of allogeneic BM transplantation shares important features typical of nonmyleoablative engraftment in rodents including durable mixed chimerism, systemic deletional donor-specific tolerance, and T-cell mediated immune resistance. This model is thus attractive for further genetic studies.
Trait variance in BM engraftment resistance
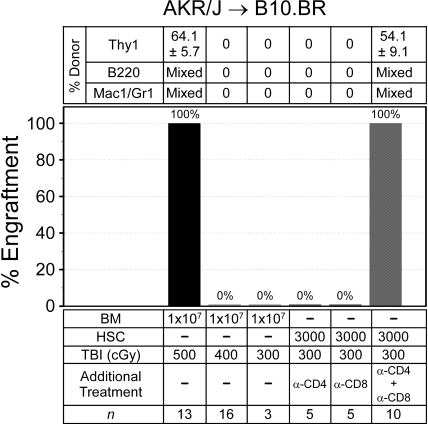
The published data describing trait variability in BM engraftment resistance is limited.29,30 Among these was work from our laboratory, where we reported that BALB.K and B10.BR mice evaluated by an HSC radioprotection assay variably resisted donor engraftment.12 Thus, we evaluated nonmyeloablative BM engraftment in AKR/J → B10.BR (H2k) mice, another MHC-matched strain combination. Recipient mice were again conditioned with a single dose of TBI and given 107 donor BM. Multilineage engraftment was evaluated in the same way as AKR/J → BALB.K mice. As shown in Figure 3, we uncovered important trait variance in engraftment barriers between B10.BR and BALB.K mice. A TBI dose of 400 cGy, which allowed BM engraftment in BALB.K mice, did not permit engraftment in B10.BR recipients. This was not reliably achieved until the TBI dose was increased to 500 cGy. The higher TBI dose in B10.BR mice at least in part reflected a requirement for overcoming immune resistance, as mice given TBI 300 cGy all engrafted with AKR/J HSCs if CD4 and CD8 T-cell depletion was added to the conditioning regimen. T cell–mediated resistance in B10.BR differed from BALB.K mice, however, as depleting either CD4 or CD8 T cells alone preserved engraftment barriers, suggesting a requirement for cooperative effects between the T-cell subsets. Trilineage mixed chimerism was confirmed in engrafted AKR → B10.BR mice for up to 4 months after transplantation.
Figure 3.
Allogeneic BM engraftment in AKR/J → B10.BR (H2k) mice. Recipient mice were given sublethal TBI, alone or with anti-CD4 (GK1.5) and/or anti-CD8 (53-6.7), before infusion of donor BM or HSCs. Shown are the percentages of mice with donor engraftment and chimerism characteristics at 6 weeks after transplantation. T-cell chimerism was evaluated by FACS for Thy1 surface markers. B-cell and myeloid chimerism was evaluated by PCR for D8Mit224 DNA markers of FACS-sorted B220+ and Mac1/Gr1+ blood cells.
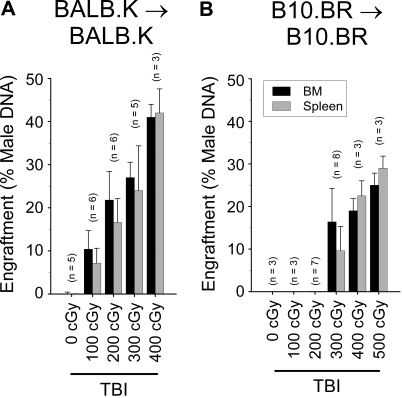
We next considered that trait variance with regard to nonimmune resistance of BM cells may contribute to the overall rejection phenotype. We tested for this possibility by conditioning BALB.K and B10.BR mice with TBI and transplanted syngeneic BM using male donors into female recipients. Because it has been shown that cell-mediated responses against Y-chromosome–encoded antigens are not elicited without presensitization,31 immune resistance in this experimental setting can be considered negligible. Engraftment was evaluated from cells harvested separately from BM and spleen of recipients 6 weeks after transplantation by quantitative PCR for male DNA. As shown in Figure 4, we again observed significant strain-specific differences. BALB.K mice engraft with syngeneic BM after as little as TBI 100 cGy conditioning, increasing to high-level engraftment as the TBI doses approached levels that also permit allogeneic engraftment (Figure 4A). In contrast, B10.BR mice strongly resist syngeneic BM engraftment, not showing detectable donor chimerism until a TBI dose of 300 cGy is given (Figure 4B). These results show that BALB.K and B10.BR mice also exhibit trait variance in nonimmune BM rejection, suggesting additional genetic control elements influencing the allogeneic BM rejection phenotype.
Figure 4.
Syngeneic BM engraftment. Syngeneic BM engraftment in (A) BALB.K and (B) B10.BR mice. Female recipient mice were given sublethal TBI and injected with syngeneic male BM at a cell dose of 107. Shown are percent donor chimerism in BM and spleen of recipient mice 6 weeks after transplantation, assayed by quantitative PCR for male-specific Sry DNA.
We attempted to account for differences in the BM engraftment phenotype by characterizing the hematologic response to TBI in these mouse strains. Ionizing radiation causes lymphocyte depletion by inducing a DNA double-strand break and inducing apoptosis.32 We harvested spleen cells from BALB.K and B10.BR mice immediately after sublethal TBI and cultured the single cell suspension in media supplemented with 20% serum for serial evaluation of apoptosis by propidium iodide staining of ethanol-fixed cells (supplemental Figure 2A). Compared with unirradiated controls, we detected high levels of apoptotic splenocytes induced within 48 hours of TBI but no difference between responses by BALB.K and B10.BR cells. We next harvested spleen, lymph node, thymus, and bone marrow from mice 72 hours after sublethal TBI and evaluated cell viability by trypan blue exclusion (supplemental Figure 2B). Compared with unirradiated controls, TBI doses permissive for allogeneic BM engraftment caused a greater than 99% reduction in viable cell numbers in all tissue compartments, but again with no significant differences between BALB.K and B10.BR mice. We analyzed the cells from the thymus by FACS for expression of CD4 and CD8 surface markers (supplemental Figure 2C). Progressive and near-total elimination of CD4/CD8 double-positive thymocytes occurred with increasing TBI doses, but in dose-response relationships that were indistinguishable comparing BALB.K and B10.BR mice. A similar FACS analysis for cKit+Lin− cells in BM after TBI revealed kinetics of depletion in this BM progenitor cell compartment that also did not differ between BALB.K and B10.BR mice (data not shown).
Collectively, these results show that BALB.K and B10.BR mice show important trait variance with regard to both immune resistance and nonimmune rejection of hematopoietic cells. This is expressed as different TBI dose requirements for engraftment, respectively, of allogeneic and syngeneic BM. Further, analyzing hematologic effects of conditioning with TBI revealed no easily detectable differences between the 2 mouse strains, suggesting that developing reasonable candidate gene approaches would be difficult.
Linkage analysis for allogeneic BM engraftment QTL
Having characterized the BM engraftment phenotypes, we proceeded with our genome-wide scan for linkage analysis. We began by generating (BALB.K × B10.BR)F1 littermates to assess whether BM engraftment results from dominance or additive effects and to determine the directionality of the allele effect. As shown in Figure 5A, we evaluated engraftment of AKR/J BM and found that F1 mice shared engraftment susceptibility identical to the BALB.K parental strain. Long-term trilineage engraftment was confirmed in all F1 mice engrafted with AKR/J BM. T-cell chimerism was evaluated by FACS analysis (Figure 5B,D). B-cell and myeloid chimerism was evaluated by RT-PCR combined with high resolution melt curve analysis of donor SNP alleles in Cd22 and Ncf2 gene transcripts (Figure 5C,E). We next transplanted syngeneic male BM into female F1 recipients and found a pattern of engraftment that again mirrored the susceptible BALB.K background (Figure 5F). These results show that engraftment, rather than rejection, of both allogeneic and syngeneic BM is a dominant allele effect.
Figure 5.
Allogeneic and syngeneic BM engraftment in (BALB.K × B10.BR)F1 mice. (A) F1 mice were given sublethal TBI and infused with AKR/J BM. Shown are the numbers of mice engrafting at 6 weeks after transplantation. For F1 mice engrafting with AKR/J BM, representative donor chimerism at 8 weeks after transplantation is shown with: (B) representative FACS analysis for T cells using Thy1.1 (donor) and Thy1.2 (host) cell-surface markers; (C) representative B-cell and myeloid chimerism by RT-PCR and melt curve analysis for Cd22 and Ncf2 coding SNP using total RNA extracted from the spleen of transplanted F1 recipient (solid line) or control mice (dotted and dashed lines); and dot plots summarizing (D) T-cell chimerism at indicated time points and (E) B-cell and myeloid chimerism at 4 months after transplantation. (F) Female F1 mice were given sublethal TBI and injected with syngeneic male BM. Shown are percent donor in BM (■) and spleen ( ) 6 weeks after transplantation by PCR for Sry DNA.
) 6 weeks after transplantation by PCR for Sry DNA.
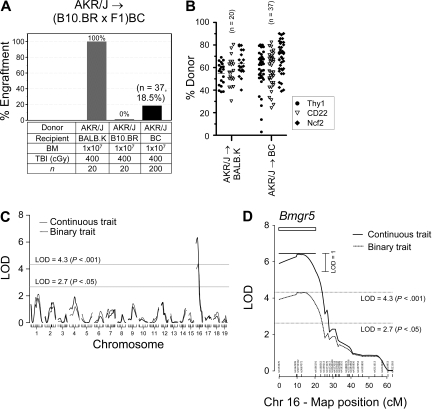
We next backcrossed F1 mice to the resistant B10.BR parental strain to generate a segregating cross for linkage analysis. We bred 200 BC mice that we maintained as sex-matched littermates before determination of the BM engraftment phenotype in each progeny. The allogeneic BM engraftment phenotype for the BC mice is shown in Figure 6A. The mice were divided into 10 groups for conditioning with TBI at a dose of 400 cGy followed by injections of AKR/J BM, each performed as individual experiments with BALB.K and B10.BR parental controls. Engraftment was evaluated at 2 separate time points, 6 weeks and again at 8 weeks after transplantation. Engraftment in control susceptible BALB.K and resistant B10.BR mice confirmed that the TBI 400 cGy dose effectively discriminated between these strains. Thirty-seven (18.5%) BC mice were scored as achieving engraftment, as defined by the presence of donor chimerism at both 6 weeks and 8 weeks after transplantation. In clinical allogeneic hematopoietic cell transplantation, donor T-cell chimerism is the best correlate of engraftment33 and, for the purpose of our studies, could be used as a continuous trait value for linkage analysis. We thus quantitated the percentage donor T-cell chimerism that was determined by FACS at 8 weeks after transplantation. As shown in Figure 6B, the median donor T-cell chimerism in the 37 BC mice with engraftment was 62.6% (range, 3.0-80.2). Not diagrammed in the plot are 163 nonengrafting BC mice, who had a median donor T-cell chimerism of 0.0% (range, 0.0-0.0). Donor engraftment in B-cell and myeloid lineage populations was confirmed in all engrafting mice by the RT-PCR and melt curve assay.
Figure 6.
Allogeneic BM engraftment QTL. The BM engraftment phenotype was assessed in [B10.BR × (BALB.K × B10.BR)F1] BC mice (n = 200) given sublethal TBI and AKR/J BM. Shown are (A) the percentage of mice with engraftment 6 and 8 weeks after transplantation and (B) the percentage of donor T cells in blood of mice that engrafted, as determined by FACS for Thy1 markers, and donor B cells and myeloid cells, as determined by RT-PCR for CD22- and Ncf2 specific SNP markers. (C) Interval mapping using R/qtl for BM engraftment considered as a continuous trait (percentage donor T-cell value; solid line) or binary trait (yes, n = 37, or no, n = 163; dotted line) plotted according to chromosome position. (D) Interval mapping of chromosome 16. The rectangular box indicates the 1 LOD confidence interval for the continuous trait and demarcates the new BM engraftment QTL termed Bmgr5. The vertical lines on the x-axis indicate positions of SNP markers used for genotyping. The horizontal dashed lines indicate significant (P < .05) and highly significant (P < .001) LOD threshold levels determined by permutation tests.
Individual BC mice were numbered and DNA was isolated from tail tip sections, before transplantation, for a 127 SNP marker genome scan. The genotype distribution in the BC mice was 49.5% B10.BR homozygous and 50.5% B10.BR/BALB.K heterozygous, not significantly different from the expected 50:50 distribution. Genome-wide linkage analysis for BM engraftment was then performed in R/qtl by simple interval mapping. We first analyzed for the engraftment phenotype as a quantitative trait using the percentage donor T-cell value as the continuous trait data. As shown in Figure 6C (solid line), a single highly significant allogeneic BM engraftment susceptibility QTL on chromosome 16 was identified. We next carried out a form of composite interval mapping by using SNP rs3667072 genotype, the individual marker with the highest LOD score, as additive and interactive covariates. We again found no evidence for linkage outside of the chromosome 16 locus (data not shown), thus evidence for a second QTL or epistatic gene effects could not be detected with the current backcross. Lastly, interval mapping using recipient gender as additive and interactive covariates revealed, respectively, no differences in BM engraftment between the sexes and no sex-specific engraftment QTL (data not shown).
Because the majority of BC mice did not engraft and had 0% as the quantitative trait value, the phenotype distribution deviated significantly from the standard assumption of normal distribution for interval mapping.20 Spikes in distributions are common in many experimental phenotypes of interest. These spikes may result from truncation by right-censored survival data or skewing due to a high amount of zero count values. This can cause a loss of power to detect QTL,34 or produce spurious linkage peaks in regions of low genotype information.35 We therefore repeated interval mapping for the engraftment phenotype as a binary trait: nonengrafting mice were scored as 0 (n = 163) and engrafting mice as 1 (n = 37). As shown again in Figure 6C (dashed line), similar results were obtained: a single highly significant locus on chromosome 16. Lastly, we applied the 2-part parametric single QTL model described by Broman for mapping QTL in the case of a spike in the phenotype distribution.36 This approach combines the analysis of the binary trait with the conditional analysis of the quantitative trait only among individuals with positive values. The chromosome 16 locus was again identified as the only significantly linked QTL (supplemental Figure 3).
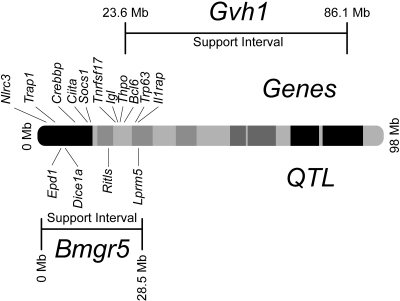
We genotyped all BC mice for 15 additional markers along chromosome 16 and repeated interval mapping. Results are shown in Figure 6D. For linkage analysis of the quantitative trait (solid line), we observed a peak LOD score of 6.4 at map position 11.1 cM, immediately flanked by SNP markers rs50732322 and rs3667072. Downstream of this peak, the linkage pattern dropped sharply and fell below the significant threshold beginning with SNP rs4172417. We used the 1 LOD confidence interval for the quantitative trait score, extending from 0.0 cM to 21.1 cM, as boundaries for a new allogeneic BM engraftment QTL. The designation Bmgr1-4 had been previously used for NK cell-mediated BM rejection QTL that mapped to chromosomes 2, 4, and 8.30 We therefore termed our chromosome 16 QTL Bmgr5. The Bmgr5 QTL interval corresponds to a segment from 0.0 to 28.5 Mb on the physical map (Figure 7). This is a gene-dense region that, as of the most recent genome assembly NCBI Build 37.1,37 contains 240 named genes, including those of the mouse immunoglobulin lambda light chain gene complex, 75 expressed sequences, 24 cDNA clones, 17 other unannotated features, and 5 uncharacterized micro-RNA.
Figure 7.
Chromosome 16 physical map. Shown are named genes and QTL mapped within the Bmgr5 confidence interval from 0.0 to 28.5 Mb, and the confidence interval for the adjacent graft-versus-host disease susceptibility locus Gvh1.
We estimated the odds ratio (OR) for BM engraftment in transplanted BC mice with a linear regression model using rs3667072 genotype as a surrogate for Bmgr5 allele. As shown in Table 2, heterozygosity at Bmgr5 increased the likelihood of achieving any donor T-cell engraftment by 5-fold (OR 5.5; 95% CI, 2.4-12.4), and for achieving engraftment with > 50% donor T-cell chimerism by 16-fold (OR 16.5; 95% CI, 4.8-56.6). Collectively, these results show that a novel chromosome 16 QTL, Bmgr5, encodes dominant susceptibility alleles from the BALB.K genetic background that has a major effect on promoting allogeneic BM engraftment in nonmyeloablated mice.
Table 2.
Regression analysis of Bmgr5 allele and bone marrow engraftment
| Engraftment phenotype* | BC mice, n | BC mice engrafting, n | Bmgr5 allele (rs3667072 genotype) | Odds ratio (95% CI) | P |
|---|---|---|---|---|---|
| Donor T-cell > 0%† | 113 | 9 | B10/B10 | 1 | |
| 87 | 28 | B10/BALB | 5.5 (2.4-12.4) | < .001 | |
| Donor T-cell > 50%‡ | 113 | 3 | B10/B10 | 1 | |
| 87 | 27 | B10/BALB | 16.5 (4.8-56.6) | < .001 |
BC indicates backcrossed; CI, confidence interval; B10, B10.BR; and BALB, BALB.K.
At 8 weeks after transplantation.
Total number of BC mice achieving bone marrow engraftment with donor T-cell chimerism > 0% (n = 37).
Total number of BC mice achieving bone marrow engraftment with donor T-cell chimerism > 50% (n = 30).
Discussion
In this report we introduce a model of nonmyeloablative hematopoietic cell transplantation using MHC-matched AKR/J donor and BALB.K and B10.BR recipient mice. Our model is notable in several regards. We had previously shown these mice to be MHC identical at the nucleotide sequence level.12 This ensured that attempts to map BM engraftment QTL would not be confounded by strong allele effects of potential MHC mismatch. At the same time, our strain combinations have more resemblance to donor-recipient pairs in clinical transplantation than those used in the prior study that identified BM engraftment QTL Bmgr1-4, which, as of this writing, is the only other published genetic analysis of BM engraftment resistance.30 That study used a cross between perforin-deficient C57BL/6 and 129/Sv mice and evaluated linkage to rejection of TAP-1 knockout BM. Our experiments in MHC-matched mice required that we develop new assays to detect long-term donor chimerism, the most stringent criteria for hematopoietic engraftment. We used these assays to characterize allogeneic and syngeneic BM engraftment, allowing BM engraftment resistance to be partitioned into 2 intermediate phenotypes, which are the immune-mediated resistance and nonimmune rejection of allogeneic hematopoietic cells. We discovered important strain-specific variability with regard to both engraftment traits, providing compelling evidence that both are under genetic control. While this singular feature of our model makes it attractive for genetic studies, equally noteworthy is our demonstration of how chimerism in our mice shares features typical of hematopoietic engraftment in nonmyeloablated rodents. This improves the likelihood that insights gained from genetic interrogation into mechanisms of specific tolerance and donor chimerism, and therefore of engraftment resistance when studied from the perspective of the recipient, using this model will have high clinical relevancy for human studies.
Results of our studies identified the Bmgr5 QTL as a novel regulator of allogeneic BM engraftment. Our genetic analysis, however, was based on a phenotypic screen for engraftment of AKR/J BM into a segregating cross bred from susceptible BALB.K and resistant B10.BR parental mice. As a result, we cannot distinguish whether Bmgr5 allele effects are mediated through immune or nonimmune resistance mechanisms. Characterization of these intermediate BM engraftment phenotypes in (BALB.K × B10.BR)F1 hybrids suggests that both mechanisms of gene action are possible. Our hypothesis of Bmgr5 gene function thus remains broad in that respect. Bmgr5 gene polymorphisms may regulate allogeneic BM engraftment by encoding an immunodominant minor histocompatibility antigen, causing graft rejection by activating T-cell responses.11 Alternatively, it could modify cellular immune responses by modifying T-cell function, analogous to polymorphisms of many immune regulatory molecules that have been implicated in graft-versus-host disease.38 Lastly, Bmgr5 genes may influence rejection of donor cells by regulating nonimmune mechanisms, such as those that mediate HSC-niche interactions (reviewed by Papayannopoulou and Scadden39), the molecular basis of which have only very recently begun to be characterized.40
In the context of these potential Bmgr5 candidate gene characteristics, we note that the confidence interval (Figure 7B) contains suppressor of cytokine signaling 1 (Socs1), a gene that encodes a protein that negatively regulates adaptive and innate immune cell activation by acting as a negative feedback loop to cytokine signaling.41 The class II transactivator (Ciita) is a master regulator of MHC class II gene expression and thus may influence immune responses through transcriptional regulation of antigen presentation.42 Bmgr5 gene function may also reflect control of responses to ionizing radiation, delivered as TBI for recipient conditioning in our model, and the p53 family member Trp63 may be considered in that regard. Prioritization of these and other candidate genes await refinement of the Bmgr5 interval by fine mapping and other approaches.
In regard to these approaches, recent advances in mouse genetics and genomic resources continue to raise prospects for accelerated positional cloning of disease susceptibility QTL. Fifteen inbred mouse strains have undergone array-based resequencing for SNP discovery.43 Because of the mosaic nature of mouse SNP haplotype blocks, this has made possible imputation of high-density SNP resources for an expanded number of laboratory mice, including all strains used in the present studies.44 Structural polymorphisms such as DNA copy number variants have recently been inventoried as well,45 representing another tool for screening candidate QTL genes for testing of causality in positional cloning. Emerging techniques such as combined cross analysis, haplotype block analysis, in silico haplotype association testing, and high-throughput approaches, such as gene expression profiling, may further enhance traditional methods for QTL localization such as congenic mapping.46 These approaches and resources have contributed, for example, to positional cloning of latexin as a regulator of hematopoietic stem cell population size and the histamine receptor H1 as a mediator of T-cell responses in autoimmune disease.47,48 We anticipate that application of these techniques will lead to successful identification of Bmgr5 genes, which may be useful for overcoming allogeneic engraftment obstacles and avoidance of engraftment failure in clinical hematopoietic cell transplantation.
Acknowledgments
This work was supported in part by grants PO1CA049605 (J.A.S.) from the National Cancer Institute, R01HL087240 (J.A.S.) from the National Heart, Lung, and Blood Institute, and R01AI076652 (T.M.C.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: T.M.C. and J.A.S. designed research, analyzed data, and prepared the manuscript; A.W.T. assisted with genetic and statistical analyses; and Y.W., S.T., and K.L. performed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thai M. Cao, Blood and Marrow Transplantation Program, University of Utah, 30 North 1900 East, SOM5C402, Salt Lake City, UT 84132; e-mail: thai.cao@hsc.utah.edu.
References
- 1.Barao I, Murphy WJ. The immunobiology of natural killer cells and bone marrow allograft rejection. Biol Blood Marrow Transplant. 2003;9:727–741. doi: 10.1016/j.bbmt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman Z, Jones M, Shatry A, Komatsu M, Mammolenti M, Levy R. Cytolytic pathways used by effector cells derived from recipient naive and memory T cells and natural killer cells in resistance to allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2005;11:957–971. doi: 10.1016/j.bbmt.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Scheffold C, Scheffold YC, Cao TM, Gworek J, Shizuru JA. Cytokines and cytotoxic pathways in engraftment resistance to purified allogeneic hematopoietic stem cells. Biol Blood Marrow Transplant. 2005;11:1–12. doi: 10.1016/j.bbmt.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Baker MB, Podack ER, Levy RB. Perforin- and Fas-mediated cytotoxic pathways are not required for allogeneic resistance to bone marrow grafts in mice. Biol Blood Marrow Transplant. 1995;1:69–73. [PubMed] [Google Scholar]
- 5.Komatsu M, Mammolenti M, Jones M, Jurecic R, Sayers TJ, Levy RB. Antigen-primed CD8+ T cells can mediate resistance, preventing allogeneic marrow engraftment in the simultaneous absence of perforin-, CD95L-, TNFR1-, and TRAIL-dependent killing. Blood. 2003;101:3991–3999. doi: 10.1182/blood-2002-09-2859. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman Z, Shatry A, Deyev V, et al. Effector cells derived from host CD8 memory T cells mediate rapid resistance against minor histocompatibility antigen-mismatched allogeneic marrow grafts without participation of perforin, Fas ligand, and the simultaneous inhibition of 3 tumor necrosis factor family effector pathways. Biol Blood Marrow Transplant. 2005;11:576–586. doi: 10.1016/j.bbmt.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Quesenberry PJ, Colvin G, Abedi M. Perspective: fundamental and clinical concepts on stem cell homing and engraftment: a journey to niches and beyond. Exp Hematol. 2005;33:9–19. doi: 10.1016/j.exphem.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abiola O, Angel JM, Avner P, et al. The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afzali B, Lechler RI, Hernandez-Fuentes MP. Allorecognition and the alloresponse: clinical implications. Tissue Antigens. 2007;69:545–556. doi: 10.1111/j.1399-0039.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 12.Cao TM, Lo B, Ranheim EA, Grumet FC, Shizuru JA. Variable hematopoietic graft rejection and graft-versus-host disease in MHC-matched strains of mice. Proc Natl Acad Sci U S A. 2003;100:11571–11576. doi: 10.1073/pnas.2035077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Caggana M, Cutler TL, Ding X. Development of a real-time polymerase chain reaction-based method for the measurement of relative allelic expression and identification of CYP2A13 alleles with decreased expression in human lung. J Pharmacol Exp Ther. 2004;311:373–381. doi: 10.1124/jpet.104.069872. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery J, Wittwer CT, Palais R, Zhou L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc. 2007;2:59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 16.Shizuru JA, Weissman IL, Kernoff R, Masek M, Scheffold YC. Purified hematopoietic stem cell grafts induce tolerance to alloantigens and can mediate positive and negative T cell selection. Proc Natl Acad Sci U S A. 2000;97:9555–9560. doi: 10.1073/pnas.170279297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao TM, Durrant D, Tripathi A, et al. Stilbene derivatives that are colchicine-site microtubule inhibitors have antileukemic activity and minimal systemic toxicity. Am J Hematol. 2008;83:390–397. doi: 10.1002/ajh.21104. [DOI] [PubMed] [Google Scholar]
- 18.Shifman S, Bell JT, Copley RR, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 20.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 23.Mangin B, Goffinet B, Rebai A. Constructing confidence intervals for QTL location. Genetics. 1994;138:1301–1308. doi: 10.1093/genetics/138.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davenport C, Kumar V, Bennett M. Rapid rejection of H2k and H2k/b bone marrow cell grafts by CD8+ T cells and NK cells in irradiated mice. J Immunol. 1995;155:3742–3749. [PubMed] [Google Scholar]
- 25.Cao TM, Shizuru JA. Engraftment of purified hematopoietic stem cells across minor histocompatibility antigen disparities are associated with distinct barriers and can be facilitated by a bone marrow-derived CD8+ cell [abstract]. Blood. 2003;98:707a. [Google Scholar]
- 26.Gandy KL, Domen J, Aguila H, Weissman IL. CD8+TCR+ and CD8+TCR− cells in whole bone marrow facilitate the engraftment of hematopoietic stem cells across allogeneic barriers. Immunity. 1999;11:579–590. doi: 10.1016/s1074-7613(00)80133-8. [DOI] [PubMed] [Google Scholar]
- 27.Marrack P, Winslow GM, Choi Y, et al. The bacterial and mouse mammary tumor virus superantigens; two different families of proteins with the same functions. Immunol Rev. 1993;131:79–92. doi: 10.1111/j.1600-065x.1993.tb01531.x. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman ZF, Levy RB. MiHA reactive CD4 and CD8 T-cells effect resistance to hematopoietic engraftment after reduced intensity conditioning. Am J Transplant. 2006;6:2089–2098. doi: 10.1111/j.1600-6143.2006.01428.x. [DOI] [PubMed] [Google Scholar]
- 29.Miller SC. Genetically determined resistance to foreign bone marrow transplantation in mice: characterization of the effector cells. J Immunol. 1983;131:92–97. [PubMed] [Google Scholar]
- 30.Johansson MH, Taylor MA, Jagodic M, et al. Mapping of quantitative trait loci determining NK cell-mediated resistance to MHC class I-deficient bone marrow grafts in perforin-deficient mice. J Immunol. 2006;177:7923–7929. doi: 10.4049/jimmunol.177.11.7923. [DOI] [PubMed] [Google Scholar]
- 31.Gordon RD, Simpson E, Samelson LE. In vitro cell-mediated immune responses to the male specific(H-Y) antigen in mice. J Exp Med. 1975;142:1108–1120. doi: 10.1084/jem.142.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barnett GC, West CM, Dunning AM, et al. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer. 2009;9:134–142. doi: 10.1038/nrc2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron F, Baker JE, Storb R, et al. Kinetics of engraftment in patients with hematologic malignancies given allogeneic hematopoietic cell transplantation after nonmyeloablative conditioning. Blood. 2004;104:2254–2262. doi: 10.1182/blood-2004-04-1506. [DOI] [PubMed] [Google Scholar]
- 34.Tilquin P, Coppieters W, Elsen JM, Lantier F, Moreno C, Baret PV. Statistical power of QTL mapping methods applied to bacteria counts. Genet Res. 2001;78:303–316. doi: 10.1017/s0016672301005365. [DOI] [PubMed] [Google Scholar]
- 35.Boyartchuk VL, Broman KW, Mosher RE, D'Orazio SE, Starnbach MN, Dietrich WF. Multigenic control of Listeria monocytogenes susceptibility in mice. Nat Genet. 2001;27:259–260. doi: 10.1038/85812. [DOI] [PubMed] [Google Scholar]
- 36.Broman KW. Mapping quantitative trait loci in the case of a spike in the phenotype distribution. Genetics. 2003;163:1169–1175. doi: 10.1093/genetics/163.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullally A, Ritz J. Beyond HLA: the significance of genomic variation for allogeneic hematopoietic stem cell transplantation. Blood. 2006;109:1355–1362. doi: 10.1182/blood-2006-06-030858. [DOI] [PubMed] [Google Scholar]
- 39.Papayannopoulou T, Scadden DT. Stem-cell ecology and stem cells in motion. Blood. 2008;111:3923–3930. doi: 10.1182/blood-2007-08-078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raaijmakers MH, Scadden DT. Evolving concepts on the microenvironmental niche for hematopoietic stem cells. Curr Opin Hematol. 2008;15:301–306. doi: 10.1097/MOH.0b013e328303e14c. [DOI] [PubMed] [Google Scholar]
- 41.Dimitriou ID, Clemenza L, Scotter AJ, Chen G, Guerra FM, Rottapel R. Putting out the fire: coordinated suppression of the innate and adaptive immune systems by SOCS1 and SOCS3 proteins. Immunol Rev. 2008;224:265–283. doi: 10.1111/j.1600-065X.2008.00659.x. [DOI] [PubMed] [Google Scholar]
- 42.Krawczyk M, Reith W. Regulation of MHC class II expression, a unique regulatory system identified by the study of a primary immunodeficiency disease. Tissue Antigens. 2006;67:183–197. doi: 10.1111/j.1399-0039.2006.00557.x. [DOI] [PubMed] [Google Scholar]
- 43.Frazer KA, Eskin E, Kang HM, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 44.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19:199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 2008;180:2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Y, Jansen M, Aronow B, Geiger H, Van Zant G. The quantitative trait gene latexin influences the size of the hematopoietic stem cell population in mice. Nat Genet. 2007;39:178–188. doi: 10.1038/ng1938. [DOI] [PubMed] [Google Scholar]
- 48.Ma RZ, Gao J, Meeker ND, et al. Identification of Bphs, an autoimmune disease locus, as histamine receptor H1. Science. 2002;297:620–623. doi: 10.1126/science.1072810. [DOI] [PubMed] [Google Scholar]