Fig. 1. Sequence alignment and location of amino acid substitution mutations in the N-terminal domain of Drosophila mtDNA helicase.
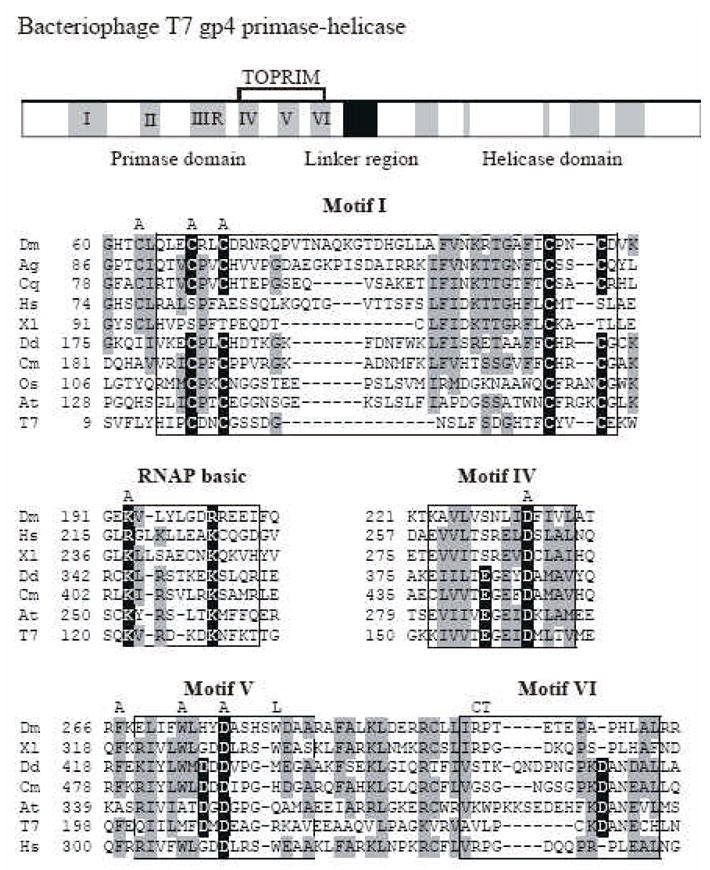
The upper panel shows a schematic diagram of the sequence organization of the bacteriophage T7 primase-helicase. The five amino acid sequence motifs common to prokaryotic primases are indicated in black numbers. Sequence alignment of relevant regions are shown below with mtDNA helicases of the fly (Dm), African malaria mosquito (Ag), Southern house mosquito (Cq), frog (Xl), man (Hs), amoeba (Dd), red alga (Cm), Rice (Os), mouse-ear cress (At) and bacteriophage T7 gp4 (T7). Data are taken from Gray et al. [6]. Amino acid residues highlighted in black are critical residues in T7 gp4, and those that are conserved in mtDNA helicases are indicated in gray. The positions of the alanine substitutions in the Drosophila mtDNA helicase used in this study are shown above the alignment as “A”. The positions of the human adPEO mutations that we analyzed in Drosophila are shown above the alignment in single letter code as “L”, “C” and “T”.
