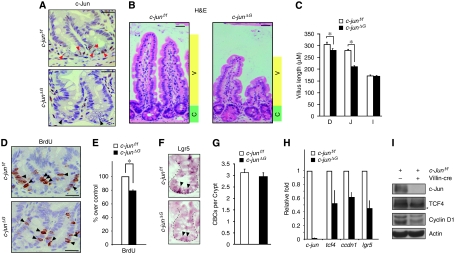
Figure 3.
Absence of c-Jun decreases crypt cell proliferation and villus length. (A) Immunohistochemistry for c-Jun on representative crypts from c-junf/f and c-junΔG intestines. Red arrow heads represent c-Jun positively labelled columnar base cells (CBC), black arrow heads represent c-Jun negative CBCs. Scale bar represents 30 μM. (B) Haematoxylin and eosin staining of jejunum epithelium from c-junf/f and c-junΔG mice. Green shading denotes proliferative zone (crypt, C) and yellow shading marks the zone of differentiation (villus, V). All animals were killed between 8–12 weeks. Scale bar represents 50 μM. (C) Quantification of the villus length from the base of the villus to the villus apex of c-junf/f and c-junΔG intestines. Histogram represents the villus length as mean ± s.e.m. in different regions of the gut (D=duodenum, J=jejunum and I=Ileum) (*P⩽0.05; student's t test). (D) Immunohistochemistry for BrdU on representative crypts from c-junf/f and c-junΔG intestines. Black arrowheads represent BrdU+ proliferative progenitors. Scale bar represents 25 μM. (E) Quantification of BrdU+ cells is represented in the histogram and expressed as % of positive cells per crypt, considering c-junf/f as 100% (*P⩽0.05; student's t test). (F) Lgr5 in situ hybridization in comparable crypts from c-junf/f and c-junΔG intestines. (G) Quantification of CBCs in c-junf/f and c-junΔG crypts. (H) qRT–PCR analysis of c-jun, ccdn1 and tcf4 transcripts in c-junΔG intestines compared with c-junf/f. The data are normalized to β-actin and represented as fold induction over c-junf/f mice. (I) Western analysis of protein lysates from c-junf/f and c-junΔG intestines for c-Jun, TCF4, cyclinD1 and β-actin (loading control). * indicates non-specific band. (n and P values are detailed in Supplementary Table S1).