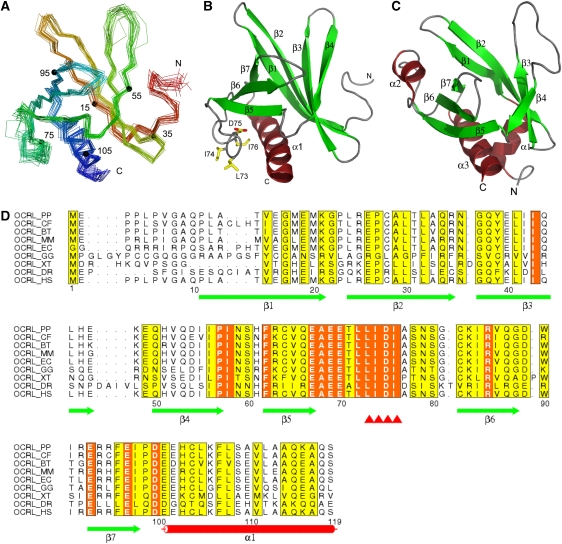
Figure 2.
NMR structure of the NH2-terminal PH domain of OCRL. (A) Superimposed Cα backbone traces of the 20 lowest-energy calculated structures. Traces are rainbow coloured with the NH2-terminus in red and the COOH-terminus in blue. Numbers and corresponding small filled circles indicate residues multiple of 20. (B) Ribbon diagram of a representative structure from (A) shown in the same orientation. Residues that are involved in clathrin binding are shown in sticks and labelled. (C) Ribbon diagram of the structure of the PH domain of PLCδ (PDB ID 1mai). (D) Sequence alignment of the NH2-terminal PH domains of OCRL orthologs. Secondary elements are drawn under the alignment. The clathrin-biding motif is marked by red triangles. This motif, as well as the preceding leucine that partially contributes to binding (see Figure 1C) is completely conserved among all OCRL orthologs. Entrez database accession numbers are as follow: OCRL_PP, GI: 55729733; OCRL_CF, GI: 74008389; OCRL_BT, GI: 156121029; OCRL_MM, GI: 45768389; OCRL_EC, GI: 149745642; OCRL_GG, GI: 118089331; OCRL_XT, GI: 115529017; OCRL_DR, GI: 118150544; OCRL_HS, GI: 13325072. PP, Pongo pygmaeus; CF, Canis familiaris; BT, Bos Taurus; MM, Mus musculus; EC, Equus caballus; GG, Gallus gallus; XT, Xenopus tropicalis; DR, Danio rerio; HS, Homo sapiens.