Abstract
Recent advances in chromatin biology have enhanced our understanding of gene regulation. It is now widely appreciated that gene regulation is dependent upon post-translational modifications to the histones which package genes in the nucleus of cells. Active genes are known to be associated with acetylation of histones (H3ac) and trimethylation of lysine 4 in histone H3 (H3K4me3). Using chromatin immunoprecipitation (ChIP), we examined histone modifications at the myosin heavy chain (MHC) genes expressed in fast vs. slow fiber-type skeletal muscle, and in a model of muscle unloading, which results in a shift to fast MHC gene expression in slow muscles. Both H3ac and H3K4me3 varied directly with the transcriptional activity of the MHC genes in fast fiber-type plantaris and slow fiber-type soleus. During MHC transitions with muscle unloading, histone H3 at the type I MHC becomes de-acetylated in correspondence with down-regulation of that gene, while upregulation of the fast type IIx and IIb MHCs occurs in conjunction with enhanced H3ac in those MHCs. Enrichment of H3K4me3 is also increased at the type IIx and IIb MHCs when these genes are induced with muscle unloading. Downregulation of IIa MHC, however, was not associated with corresponding loss of H3ac or H3K4me3. These observations demonstrate the feasibility of using the ChIP assay to understand the native chromatin environment in adult skeletal muscle, and also suggest that the transcriptional state of types I, IIx and IIb MHC genes are sensitive to histone modifications both in different muscle fiber-types and in response to altered loading states.
Keywords: hindlimb suspension, chromatin immunoprecipitation, in vivo, multigene family
muscle contraction provides the basis for all movements and postural support in animals and humans. A broad range of contractile intensities can exist in different muscle fibers, primarily attributed to diversity in the motor protein myosin heavy chain (MHC). Four isoforms of MHC (I, IIa, IIx, IIb), each encoded by a distinct gene, can be expressed in adult skeletal muscle. Intrinsic differences in the ATPase (and shortening velocity) properties of the MHC isoforms have led to the classification of slow and fast fiber-types in muscle (5). For example, slow type I MHC predominates in the soleus, whereas fast IIx and IIb are the primary MHCs expressed in the plantaris. The transcriptional products of the MHC genes are also easily altered by perturbations to environmental stimuli, such as altered mechanical loading (5, 16). DNA regulatory sequence, transcription factors, and other mechanisms that are important in the transcriptional regulation of the MHC genes in skeletal muscle have been studied; however, virtually no information is known about the role of chromatin remodeling at these highly regulated genes.
Recent advances in the understanding of the dynamic nature of DNA regulatory regions have highlighted the importance of chromatin structure and associated histone modifications in influencing gene expression. Numerous chromatin-bound protein complexes and their enzymatic activities have been identified, which can covalently modify certain histone sites that, in turn, determine whether a specific gene will be transcribed or not (28). Posttranslation modifications such as acetylation, methylation, and phosphorylation to histone tails can exert control on a given gene's expression. The combination, sequence and extent of various histone modifications, referred to as the histone/epigenetic code hypothesis, is proposed to result in distinct functional consequences impacting various cellular processes (35, 42). Of particular relevance in understanding the regulation of the MHC genes is that these histone modifications are predicted to ultimately result in varying degrees of transcriptional activation or repression.
Histone hyperacetylation is correlated positively with actively transcribed genes (2, 22, 38). Acetylation alters the charge of histone tails, and thus interaction properties of both histone-DNA contacts within a given nucleosome, and also internucleosomal contacts to result in relaxing of a compact chromatin structure (10, 35) This enables access of compacted DNA to different factors that promote transcription of the coded gene. The activation of muscle-specific genes during myofiber differentiation is dependent on the activities of enzymes that regulate histone acetylation, i.e., histone acetyltransferases (HATs) and histone deacetylases (HDACs) (26). Myogenic basic helix-loop-helix (bHLH) MyoD, Myf5, myogenin, and Mrf4 proteins and members of the MEF2 family of MADS-box transcription factors bind to the regulatory regions of muscle-specific genes to activate them (3, 7, 26, 37). These muscle-specific proteins are reported to activate and repress muscle-specific genes by associating with HATs and HDACs, respectively, to control the acetylation state of histones during differentiation (26, 37), and in an activity-dependent manner in adult muscle (27, 44).
Several recent reports have provided evidence that indicate that particular HDAC isoforms may act as nodal points of control for skeletal muscle fiber-type phenotype in vivo by mediating, via muscle-specific transcription factors, the expression of genes involved in the neuromuscular junction, metabolism (glycolytic vs. oxidative), and contraction (slow vs. fast) (20, 33, 43, 44). A critical role for HDACs in the coordinated regulation of MHC genes has been demonstrated with various manipulations to HDAC isoforms in both skeletal (20, 33, 44) and cardiac muscle (21, 25). It is not clear where the effects of manipulations to HDACs, which oppose the enzymatic activity of HATs, are mediated. Such effects could be mediated at the level of signaling intermediates, at the level of transcription factors (e.g., MEF2, myogenin) that bind the promoters of MHC genes, at the level of the histones of the MHC genes themselves, or at any combination of the above. Furthermore, the acetylation state of signaling proteins and transcription factors themselves may be altered in addition to the histones at their promoters (41). In the present study we initiate a series of experiments to examine the acetylation state of histone H3 (H3ac) of the MHC genes themselves.
In addition to H3ac, we examined trimethylation of histone H3 at lysine 4 (H3K4me3). This histone modification also is associated with active gene transcription (6, 28, 32, 36). Although the precise role for H3K4me3 marks has not been fully characterized, H3K4me3 has been shown to be coupled to transcription through the recruitment of effectors, which are subunits of chromatin remodeling complexes that are required for transcriptional competence (34). Recruitment of effector proteins via H3K4me3 may also stabilize the association of sequence-specific trans-acting factors/complexes with the chromatin template, and provide the cell with a gene-specific regulated response (34). A high correlation is observed in genome-wide analyses between H3ac and H3K4me3 within the 5′-regions of active genes (13, 38, 48). Recent reports have further demonstrated that H3K4me3 may be a modification that serves as a target for HATs and HDACs (9, 15). Thus, genomic regions trimethylated at H3K4 may be subjected to the opposing actions of acetylation and deacetylation in a dynamic manner, such that turnover of acetyl groups is high, thereby ensuring a rapid transcriptional response to altered conditions.
Since the histone tail modifications H3ac and H3K4me3 have both been shown to occur at active genes, and may be functionally interconnected with each other to recruit and stabilize transcription complexes, we chose to focus on these two modifications in the context of the four MHC genes that have varying levels of transcription among its members, and between muscles of different type as well as different environments. To our knowledge, a survey of histone modifications at MHC genes in skeletal muscle has not been carried out. Therefore, we sought to determine whether H3ac and H3K4me3 are differentially enriched at MHC promoters in vivo in the following conditions: 1) under steady-state transcriptional states where the MHC genes in different fiber types have a stable yet divergent MHC transcription patterns, and 2) in response to altered mechanical loading states, where transcription of the MHC genes become dynamically altered.
We report that both H3ac and H3K4me3 enrichment at each of the four MHC genes corresponds with transcript abundance in the fast fiber-type plantaris (Pla) and slow fiber-type soleus (Sol). A slow to fast fiber type shift in the Sol (i.e., with unloading or unloading plus thyroid hormone) similarly induced alterations to H3ac and H3K4me3 at the type I, IIx, and IIb MHC genes, but not the IIa MHC.
METHODS
Animal procedures.
Female Sprague-Dawley rats (140–150 g) were used for all experiments. Animals were randomly assigned to either control or hindlimb suspension (HS) groups (n = 7/group). Control animals were housed in groups of four in a temperature- and light-controlled environment (i.e., 12:12 h light-dark cycle). All animals in a given experiment were allowed food and water ad libitum, and all procedures were approved by the Institutional Animal Care and Use Committee. HS was carried out for 7 days, which was shown in prior experiments to be sufficient to induce measurable alterations in the endogenous MHC genes expression (authors' unpublished observations). Animals subjected to thyroid hormone treatment were administered 150 μg·kg−1·day−1 of triiodothyronine (T3) by intraperitoneal injection. At the end of the experiment, rats were euthanized and the muscles were rapidly removed, weighed, and frozen at −80°C for later analysis.
Hindlimb suspension protocol.
The HS model used employed a tail traction method using a noninvasive tail casting procedure described previously (46). The technique used a swivel harness system incorporated into the casting materials, which was attached to a hook at the top of the cage. The hook was adjusted to allow only the forelimbs of the animal to reach the floor of the cage. Suspended animals were free to move about the cage using their forelimbs to obtain food and water.
RNA analysis.
Total RNA was extracted from frozen control plantaris (Pla), control soleus (Sol), and from HS soleus (HS Sol) using the Tri Reagent protocol (Molecular Research Center). Extracted RNA was DNase-treated using one unit of RQ1 RNase-free DNase (Promega) per microgram of total RNA and was incubated at 37°C for 30 min followed by a second RNA extraction using Tri Reagent LS (MRC).
RT-PCR was used to assess pre-mRNA and mRNA of target genes. RT-PCR reactions were performed with the OneStep RT-PCR Kit (Qiagen), where the RT and PCR are performed in a single reaction tube, with some modifications to the manufacturer's protocol, and as described previously (31). This protocol has been optimized to avoid amplification of nonspecific transcripts, which are known to be coamplified with pre-mRNA and mRNA transcripts, and can thus preclude accurate measurement (14, 31). These one-step RT-PCR analyses were performed using 10 ng to 200 ng total RNA and 15 pmol of specific primers in 25-μl total volume and were carried out on a Robocycler (Stratagene). Samples to be compared were run under similar conditions (template amounts, PCR cycle numbers). RT reactions were performed at 50°C for 30 min followed by 15 min of heating at 95°C, followed by PCR cycling for a varied number of cycles (20–32 cycles). The annealing temperature was based on the PCR primers optimal annealing temperature. PCR primers used for RNA analysis are shown in Table 1. The amount of RNA and the number of PCR cycles were adjusted so that the accumulated product was in the linear range of the exponential curve of the PCR amplifications. PCR products were separated by electrophoresis on agarose gels and stained with ethidium bromide. The ultraviolet light-induced fluorescence of stained DNA was captured by a digital camera, and band intensities were quantified by densitometry with ImageQuant software (Molecular Dynamics) on digitized images.
Table 1.
PCR primer sequences, their specific target, and PCR product size, for primers used for RNA analysis of MHC genes
| Target | RT-PCR Primers: 5′→3′ | PCR Product Size, bp |
|---|---|---|
| Type I pre-RNA | Forward: CCTGGTCCTATGTGCCGATCTCTAACGA | 215 |
| Reverse: CGGTCCCCAATGGCAGCAATAAC | ||
| Type I mRNA | Forward: GGAGCTCACCTACCAGACAGA | 308 |
| Reverse: CTCAGGGCTTCACAGGCATCC | ||
| IIa pre-RNA | Forward: TGCTTCCCAATGCTGCCATATCTACAT | 295 |
| Reverse: TTCCTACTGCTTCCCTTGGTCTTGTCA | ||
| IIa mRNA | Forward: CCTCTTACTTCCCAGCTGCACCTTCT | 239 |
| Reverse: ACTTTCCCTGCGTCTTTGCTCTGAAT | ||
| IIx pre-RNA | Forward: TGCCACAGAAAGAGGGACGC | 290 |
| Reverse: CTGGCTGTGGTGTGGCTGAAA | ||
| IIx mRNA | Forward: ACGGTCGAAGTTGCATCCCTAAAG | 263 |
| Reverse: CACCTTCGGTCTTGGCTGTCAC | ||
| IIb pre-RNA | Forward: GGCCATGCCAGCTAGCTTTTACG | 270 |
| Reverse: GCGTTTTGATTGGTGGAAGAGTCC | ||
| IIb mRNA | Forward: AGCCTGCCTCCTTCTTCATCTGG | 229 |
| Reverse: CACGGTTGCTTTCACATAGGACTC | ||
| β-Actin | Forward: CACGCCCTTTCTCAATTGTCTTTCT | 225 |
| Reverse: GGCCATTTATCACCAGCCTCATTAG |
MHC, myosin heavy chain.
Posttranscriptional control of mRNA steady-state levels can occur at many steps after the synthesis of the initial pre-mRNA and is subject to stability regulation (10). The pre-mRNA transcript abundance serves as a better marker of a gene's level of transcriptional activity than the mRNA because its half-life is much shorter. The nuclear run-on assay is another method to quantitate a gene's transcriptional activity; however, in our hands it is a technically unreliable measurement tool for the MHC genes, due to the inability to detect outcomes with consistent fidelity. Moreover, assessing the transcriptional activity of other genes by measuring pre-mRNA with RT-PCR has been validated as an alternative to the nuclear run-on approach (12).
MHC mRNA isoform distribution.
The MHC mRNA isoform distribution was evaluated by RT with oligo(dT)/random primers followed by PCR with primers targeting the embryonic, neonatal, I, IIa, IIx, and IIb MHC mRNAs, as described previously (11, 49). In these PCR reactions, each MHC mRNA signal was corrected to an externally added control DNA fragment that was coamplified with the MHC cDNAs using the same PCR primer pair. This approach provides a means to correct for any differences in the efficiency and/or pipetting of each PCR reaction. A correction factor was used for each control fragment band on the ethidium bromide-stained gel to account for the staining intensity of the variably sized fragments (224 to 324 bp), as reported previously (11).
Tissue preparation.
Frozen muscle tissue was minced, and then washed in ice-cold PBS. All solutions were supplemented with protease inhibitors (leupeptin, AEBSF, and aprotinin, each at 1:1,000). Minced tissue was then incubated for 10 min in 1% formaldehyde to cross-link chromatin-DNA. Cross-linking was stopped by addition of glycine to 0.125 M for 5 min. This solution was exchanged with cold PBS, and then repeated a second time to remove all the formaldehyde. Tissue samples were then homogenized in PBS (20 vol of the muscle weight) with a Dounce homogenizer. The homogenate was then pelleted by centrifugation at 1,500 g for 10 min. The pelleted muscle tissue was resuspended in cold SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.1), and sonicated (model VCX 130, Sonics Vibracell) to fragment the DNA. Samples were centrifuged at 12,000 g for 10 min to remove insoluble material. To ensure effectiveness of sonication, an aliquot of the supernatant was reverse cross-linked by incubation at 65°C overnight, and RNase treated (RNase A). Then the protein was digested (proteinase K) and then run on a 2% agarose gel to confirm size of DNA fragments to be between 200 and 1,000 bp. This aliquot was also used to measure the DNA concentration of the chromatin-DNA. This is necessary because of the varied muscle mass to DNA ratio in the three types of muscle samples analyzed. HS Sol tissue is more enriched in DNA than Sol, and Sol has more DNA than Pla per unit muscle mass. Therefore, to equalize the starting DNA concentrations for the chromatin immunoprecipitation (ChIP) assay, we used SYBR green I to bind DNA. A Stratagene Mx3000p real-time PCR machine was used in the quantitative plate read mode to accurately measure DNA concentration, with thymus calf DNA (Sigma) used as a standard.
Chromatin immunoprecipitation.
Approximately 25 μg of DNA of each muscle sample was used to perform chromatin immunoprecipitation. Normal rabbit IgG (12-370) and anti- H3ac (06-599) was obtained from Millipore (Billerica, MA). The H3ac antibody detects diacetylation at lysines 9 and 14. Anti-histone H3 (ab1791) and anti-H3K4me3 (ab8580) was obtained from Abcam (Cambridge, MA). Chromatin was precleared with protein A/G agarose (Pierce, Rockford, IL) by incubation for 30 min at 4°C on a rotating platform, in a volume of 1 ml with ChIP dilution buffer (0.01% SDS, 1.1%, Triton X-100, 1.2 mM EDTA, 16.7 mM Tris·HCl, pH 8.1, and 167 mM NaCl). After agarose removal by centrifugation, 1% of the precleared chromatin was saved and used as input DNA. Antibody was added to chromatin and incubated at 4°C with rotation for ∼1 h. Protein A/G agarose was added and incubated at 4°C with rotation for ∼2 h. Three separate washes with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris·HCl, pH 8.1, and 150 mM NaCl) were then performed, followed by separate washes for 15 min with rotation with the following buffers. High salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris·HCl, pH 8.1, and 500 mM NaCl), LiCl [0.25 M LiCl, 1% Igepal-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA, and 10 mM Tris, pH 8.1], and two separate washes with TE buffer (10 mM Tris·HCl, 1 mM EDTA, pH 8.0).
Chromatin-DNA complexes were eluted (0.1 M NaHCO3, 1% SDS) from agarose beads, and cross-links were reversed by incubation at 65°C overnight. Samples were RNase treated (RNase A), protein was digested (proteinase K), and DNA was purified by using a spin column (Qiaquick PCR purification kit). Immunoprecipitated DNA for specific genes was analyzed and quantified by PCR. PCR primers used with ChIP samples are shown in Table 2. The number of PCR cycles and amount of ChIP DNA was adjusted so that the accumulated product was in the linear range of the exponential curve of the PCR amplifications. PCR products were separated by electrophoresis on agarose gels and stained with GelGreen (Biotium). The ultraviolet light-induced fluorescence of stained DNA was captured by a digital camera, and band intensities were quantified by densitometry with ImageQuant software (Molecular Dynamics) on digitized images.
Table 2.
PCR primer sequences, their specific target, and PCR product size, for primers used for ChIP of MHC genes
| Target | RT-PCR Primers: 5′→3′ | PCR Product Size, bp |
|---|---|---|
| Type I | Forward: GGCCTGGGCCTACCTCTTTATCC | 286 |
| Reverse: TATTCAATTGGGGCACTCTTCGGGTGTAT | ||
| IIa | Forward: ATCATTACCCCAAATATCACCCTATCC | 323 |
| Reverse: GGCCCCAGATGCACATTACACTA | ||
| IIx | Forward: TGCCACAGAAAGAGGGACGC | 290 |
| Reverse: CTGGCTGTGGTGTGGCTGAAA | ||
| IIb | Forward: AGGGAATAAATGTTAACTTGTTGACACTGG | 218 |
| Reverse: GGGGGCGGGGCTAATGAAGC | ||
| β-Actin | Forward: CACGCCCTTTCTCAATTGTCTTTCT | 225 |
| Reverse: GGCCATTTATCACCAGCCTCATTAG |
ChIP, chromatin immunoprecipitation assay.
With the ChIP assay, primers can be targeted to any genomic region. The histone modifications H3ac and H3K4me3 have been shown previously to peak immediately downstream of the transcription start sites of active genes (6, 13, 51). A preliminary analysis of these histone modifications in the MHC genes resulted in the same conclusion. Thus, PCR primers were designed to target the second intron of each MHC gene studied, which occurs before the translation start site (ATG) of each gene. This location occurs at 1.1 to 1.4 kb from the transcription start site of the ∼25 kb MHC genes.
For each sample, four ChIP assays were carried out in parallel reactions with antibodies to H3ac, H3K4me3, core histone H3, and normal rabbit IgG. The latter serves as a negative control for specificity of antibody binding. For analysis, the normal rabbit IgG IP signal was subtracted from the specific antibody IP signals. In all cases, the normal IgG precipitated negligible levels of DNA for the targeted genes analyzed. Then this value was divided by the input DNA PCR signal, to correct for any differences in starting DNA concentrations between samples, and for differences in DNA accessibility at the PCR-targeted genomic sites. Previous studies have shown that variation in histone occupancy at different genomic regions, or in response to varied gene activity, can lead to misinterpretation of histone modifications measured with ChIP (32, 51). This is because nucleosome density inversely correlates with transcription rate (23). Therefore, all data are expressed relative to core histone H3 ChIP results at each PCR-targeted genomic site.
Statistical analyses.
Data are reported as means ± SE. Differences between three muscle groups (Pla, Sol, and HS Sol) were analyzed using one-way ANOVA, with Newman-Keuls post hoc test. Differences between two groups were analyzed using an unpaired t-test. Statistical significance was set at P < 0.05.
RESULTS
RNA transcripts of the MHC genes.
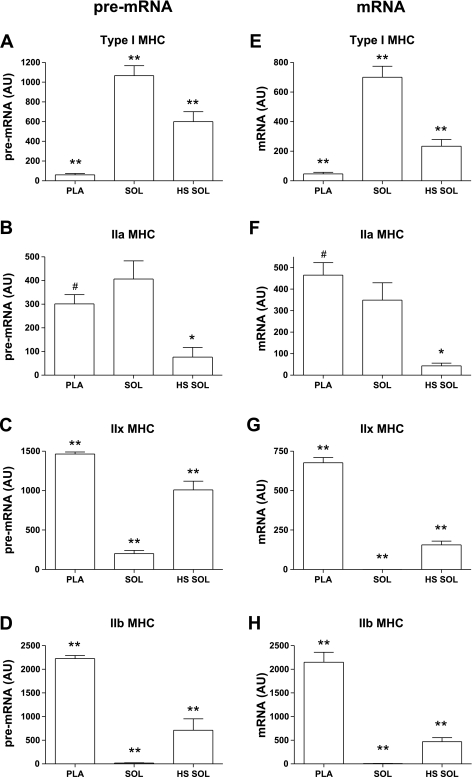
The profile of pre-mRNA and mRNA transcripts of the Pla, Sol, and HS Sol are shown in Fig. 1. The unspliced pre-mRNA products (Fig. 1, A–D) show congruency with the spliced mRNA products (Fig. 1, E–H) when comparing expression level between the three different muscle groups examined. Pre-mRNAs are the nascent, unprocessed, transcriptional products, and they serve as an indirect measure of the level of transcriptional activity of the corresponding gene.
Fig. 1.
pre-mRNA and mRNA transcripts of the myosin heavy chain (MHC) genes in plantaris (Pla), soleus (Sol), and hindlimb suspension soleus (HS Sol) muscle groups. Bar graphs show means ± SE of RT-PCR analyses. A–D: pre-mRNA. E–H: mRNA. AU, arbitrary units. *Significantly different from Sol; **all groups significantly different from each other; #significantly different from HS Sol (P < 0.05); n = 6/group.
The classification of fast and slow fiber types is illustrated by the distinct expression of MHC transcripts in the Pla and Sol (Fig. 1 and Table 3). As a percentage of total MHC, the fast fiber-type Pla expresses a predominance of IIx and IIb MHC mRNA, while the slow fiber-type Sol muscle expresses primarily the slow type I MHC mRNA, and smaller proportion of the fast IIa MHC (Table 3). Differential expression of the type I, IIx, and IIb MHC between the Pla and Sol is also observed in comparing the relative abundance of pre-mRNA and mRNA between these two muscle types (Fig. 1). Expression of IIa MHC pre- and mRNA is similar between the Pla and Sol (Fig. 1, B and F).
Table 3.
MHC mRNA isoform distribution expressed as a percentage of total MHC mRNA
| I | IIa | IIx | IIb | Embryonic | Neonatal | |
|---|---|---|---|---|---|---|
| Pla | 3% | 12% | 31% | 54% | 0% | 0% |
| Sol | 88% | 4% | 0% | 0% | 8% | 0% |
| HS Sol | 65% | 0% | 15% | 18% | 2% | 0% |
Pla, plantaris; Sol, soleus; HS, hindlimb suspension; n = 6 per group.
With HS, the Sol is remodeled to a muscle more resembling the fast Pla, where IIx and IIb MHC become strongly expressed (Fig. 1, C and D, and Table 3). Type I MHC is downregulated in HS Sol as compared with Sol, although its pre-mRNA is still transcribed in the HS Sol, indicating that the type I MHC gene remains transcriptionally active after 7 days of HS (Fig. 1A). Transcriptional products of IIa MHC are significantly reduced in HS Sol as compared with Sol (Fig. 1, B and F).
The proportion of the developmentally regulated embryonic and neonatal MHCs is also shown in Table 3. Although expression of the embryonic MHC is downregulated in HS Sol compared with Sol as a percentage of total MHC (Table 3) and at the absolute level of pre- and mRNA abundance (data not shown), we have previously shown that it is not translated into protein in adult muscle and therefore does not contribute to the functional performance of the muscle (11).
Histone H3 acetylation.
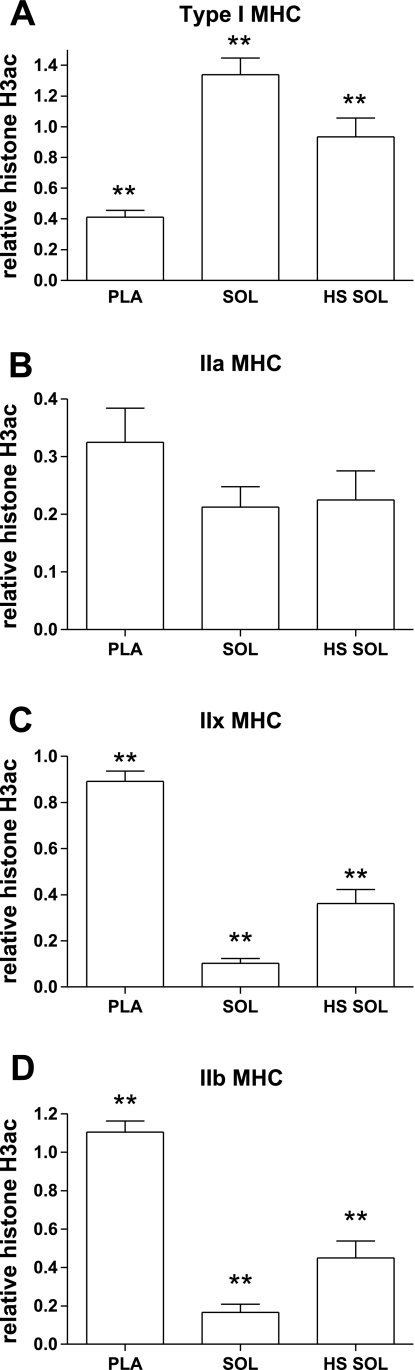
The H3ac data shown are expressed relative to the core histone H3, to account for variation in histone H3 occupancy between the different muscle samples with differing rates of gene transcription. Histone occupancy is subject to alteration depending on the transcriptional state of the gene, such that nucleosome occupancy inversely correlates with transcription rate (23). H3ac was assessed at genomic regions of the second intron of each of the MHC genes. Significant fiber-type differences in H3ac were observed at each of the MHC genes with differential transcriptional expression between the slow Sol vs. fast Pla muscles (i.e., I, IIx, and IIb; Fig. 2). The H3ac pattern in the type I, IIx, and IIb MHC genes corresponds to its MHC expression profile (compare Fig. 1 and Fig. 2). Thus, the fast Pla, which maintains an abundance of IIb and IIx MHC, was highly enriched in H3ac at both the IIb and the IIx MHC genes in contrast to the Sol. The slow Sol has relatively little expression of the fast IIx and IIb MHCs, and in correspondence to this expression the fast IIx and IIb MHCs are deacetylated relative to the Pla. A reciprocal pattern is observed in the slow type I MHC, where type I mRNA expression is low in the Pla and highly abundant in the Sol, and H3ac is similarly low in the Pla and relatively enriched in the Sol. H3ac at the IIa MHC did not differ significantly however, between the Sol and Pla, which corresponds to the lack of difference in IIa MHC transcription in the two muscle types. The directional shifts of mRNA and the corresponding H3ac among the MHCs in Pla versus Sol are summarized in Table 4.
Fig. 2.
Relative histone H3 acetylation (H3ac) at MHC genes. A: type I MHC. B: IIa MHC. C: IIx MHC. D: IIb MHC. Bar graphs show means ± SE of chromatin immunoprecipitation (ChIP) analyses; n = 7/group. Histone H3 acetylation is corrected for core histone H3 occupancy, as described in methods. **All groups significantly different from each other (P < 0.05).
Table 4.
Summary of directional changes in mRNA abundance and enrichment of H3ac and H3K4me3 in each of the four MHCs in Pla and HS Sol as compared with Sol
| MHC Gene | Variable Assessed | Pla versus Sol | HS Sol versus Sol |
|---|---|---|---|
| I | mRNA | ↓↓ | ↓ |
| H3ac | ↓↓ | ↓ | |
| H3K4me3 | ↓↓ | ⇆ | |
| IIa | mRNA | ⇆ | ↓↓ |
| H3ac | ⇆ | ⇆ | |
| H3K4me3 | ⇆ | ⇆ | |
| IIx | mRNA | ↑↑ | ↑ |
| H3ac | ↑↑ | ↑ | |
| H3K4me3 | ↑↑ | ↑ | |
| IIb | mRNA | ↑↑ | ↑ |
| H3ac | ↑↑ | ↑ | |
| H3K4me3 | ↑↑ | ↑ |
Directional arrows indicate directional change in indicated variable, relative to control Sol. ↓↓, highly decreased; ↑↑, highly increased; ↓, decreased; ↑, increased; ⇆, no change. H3ac, histone H3 acetylation; H3K4me3, trimethylation of lysine 4 on histone H3.
When an inactivity model is imposed on the slow Sol muscle, a faster MHC transcriptional pattern emerges (Fig. 1 and Table 3). The H3ac pattern was similarly altered for the type I, IIx, and IIb MHCs. H3ac is more highly enriched in the IIx and IIb MHC genes in HS Sol muscle as compared with Sol (Fig. 2, C and D, respectively). Conversely, the type I MHC became deacetylated in the HS Sol as compared with the Sol. In both of these cases, MHC transcription positively corresponds with H3ac (compare Fig. 1 and Fig. 2). However, H3ac in the IIa MHC did not correspond with its transcription pattern. Transcription of the IIa MHC is significantly decreased in the HS Sol compared with Sol, while H3ac was unchanged with HS. Thus, the directional shifts in types I, IIx and IIb pre-mRNA and mRNA in HS Sol as compared with Sol correspond to that of H3ac (see Table 4).
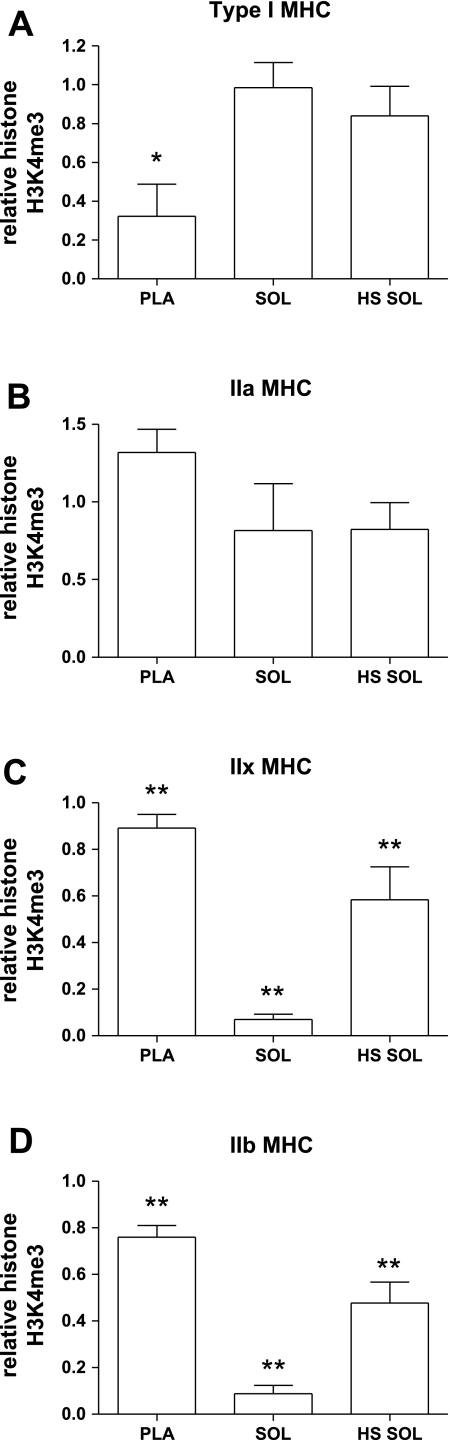
Histone H3 lysine 4 trimethylation.
Together with histone H3 acetylation, we assayed the MHC genes for H3K4me3. Similar to H3ac, H3K4me3 was highly enriched at the second intron of the IIx and IIb genes in the Pla relative to the Sol, which similarly corresponds with the transcriptional activity of these MHC genes in each muscle type (compare Fig. 1 and Fig. 3). H3K4me3 at the type I MHC also corresponds with type I transcription, such that trimethylation is increased in the Sol as compared with the Pla, where type I MHC mRNA is expressed at low levels. Upon induction of type IIb and IIx MHC transcription, which occurs upon unloading of Sol, we observed significantly increased enrichment of H3K4me3 at these genes in HS Sol as compared with Sol (Fig. 4). There was no statistically significant difference in H3K4me3 in the IIa MHC between the three muscle types studied or type I MHC between the Sol and HS Sol (Fig. 4). The directional shifts of mRNA and the corresponding H3K4me3 among the MHCs in Pla versus Sol and HS Sol versus Sol are summarized in Table 4.
Fig. 3.
Relative trimethylation of lysine 4 on histone H3 (H3K4me3) at MHC genes. A: type I MHC. B: IIa MHC. C: IIx MHC. D: IIb MHC. Bar graphs show means ± SE of ChIP analyses; n = 6/group. Histone H3K4me3 is corrected for core histone H3 occupancy, as described in methods. *Significantly different from Sol (P < 0.05). **All groups significantly different from each other (P < 0.05).
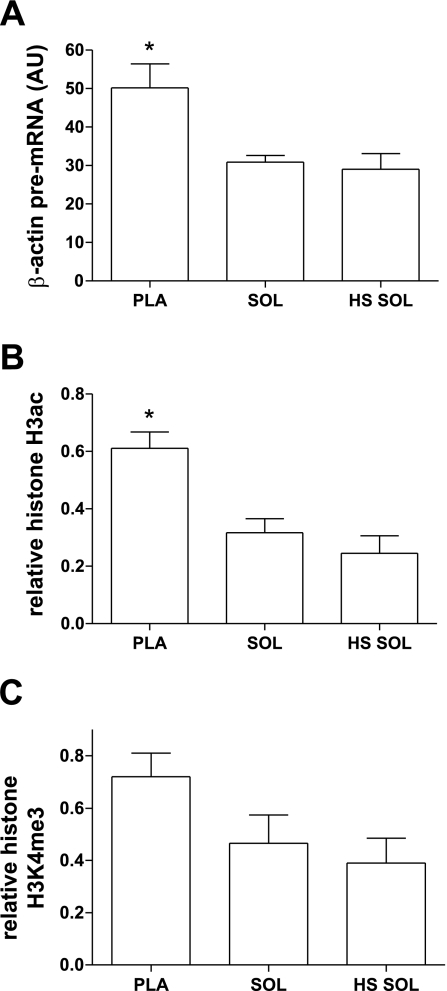
Fig. 4.
β-Actin transcription and relative modifications to H3ac and H3K4me3 in Pla, Sol, and HS Sol. A: pre-mRNA. B: histone H3ac. C: histone H3K4me3. Bar graphs show means ± SE; n = 6/group. Histone H3ac and histone H3K4me3 are corrected for core histone H3 occupancy, as described in methods. *Significantly different from Sol and HS Sol (P < 0.01).
Histone modifications at β-actin gene.
To validate the results of the ChIP assays in determining the levels of enrichment of H3ac and H3K4me3 at the MHC genes, we examined the H3ac- and H3K4me3- ChIP DNA for another target gene, β-actin, which is less prone to dynamic change, particularly in response to unloading of Sol (Fig. 4). The β-actin gene is constitutively active in skeletal muscle, and its expression is unaltered in HS Sol compared with Sol (Fig. 4A). Thus, β-actin serves as a control gene unaffected by HS. ChIP analyses showed that enrichment of H3ac and H3K4me3 at the 5′-region of the β-actin gene also does not differ between Sol and HS Sol (Fig. 4, B and C). The Pla had significantly greater β-actin pre-mRNA expression than either Sol or HS Sol, and this corresponded with the significantly increased enrichment of H3ac although not H3K4me3.
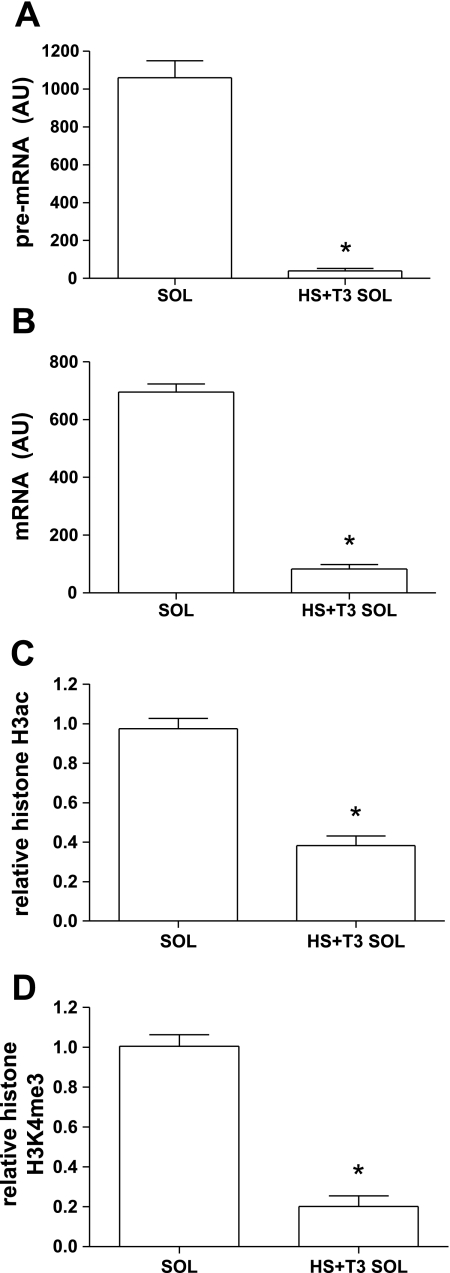
Repression of type I MHC with HS + T3.
Since there was a lack of correspondence between H3K4me3 and type I mRNA between Sol and HS Sol, we then determined whether an alternative model, in which the type I MHC could be more robustly repressed than HS alone, would result in a loss of H3K4me3 enrichment at the type I MHC promoter. In a separate experiment we found that HS combined with thyroid hormone treatment (HS + T3) results in virtually complete repression of type I MHC transcription (Fig. 5, A and B). This also resulted in a significant reduction of both H3ac (Fig. 5C) and H3K4me3 (Fig. 5D) at the type I MHC. Thus, the loss of H3K4me3 at the type I MHC with more robust repression indicates that HS alone likely resulted in insufficient repression of type I MHC to alter H3K4me3.
Fig. 5.
Type I MHC transcription and relative modifications to H3ac and H3K4me3 in Sol and HS + triiodothyronine (T3) Sol. A: pre-mRNA. B: mRNA. C: histone H3ac. D: histone H3K4me3. Bar graphs show means ± SE; n = 6/group. Histone H3ac and histone H3K4me3 are corrected for core histone H3 occupancy, as described in methods. *Significantly different from Sol (P < 0.01).
DISCUSSION
As an initial survey of histone modifications involving the chromatin of the MHC genes, we examined acetylation of histone H3 and trimethylation of H3 lysine 4 in each of the four MHCs expressed in adult rodent skeletal muscle. We compared the extent of these chromatin modifications, previously shown to positively correlate with gene activity, in three types of muscle: fast versus slow fiber-type muscles together with muscle unloading, which is shown to downregulate types I and IIa MHC and upregulate types IIx and IIb MHC. We report that enrichment of H3ac corresponds to mRNA abundance in the type I, IIx, and IIb MHCs in each of the three muscle groups (Pla, Sol, HS Sol). As the soleus myofiber is remodeled in response to unloading, downregulation of the type I MHC corresponds with deacetylation of histones, while upregulation of the IIx and IIb MHC genes occurs in concert with enhanced histone acetylation (see Table 4).
In comparing the fast fiber-type Pla with the slow fiber-type Sol, mRNA/pre-mRNA abundance of each of the four MHCs corresponds with both H3ac and H3K4me3. This comparison demonstrates that the fiber-type differences in MHC transcription are linked to modifications at the level of the chromatin of the genes that primarily define the fast vs. slow fiber phenotype. H3K4me3 is also increased in HS Sol as compared with Sol in both the type IIx and IIb MHCs, appearing in correspondence with the induction of these two genes that occurs in HS Sol. An almost identical pattern of histone acetylation is observed in the Pla to that seen in the HS Sol, when compared with the Sol, although the directional change is more exaggerated in the Pla as compared with HS Sol (summarized in Table 4). The directional difference of mRNA/pre-mRNA is similarly exaggerated in the Pla, indicating that the level of transcriptional activity of the MHCs is reflected in the degree of enrichment of epigenetic markers of gene activity.
The globin genes are another multigene family that undergoes coordinated shifts in gene expression, in liver tissue. In the fetal vs. adult stages of development, expression of the globin genes switches in correlation with the level of histone acetylation at the developmentally regulated globin genes (51). Hyperacetylation of histone H3 was observed in the active β-globin genes in adult tissue, while the inactive embryonic globin genes exist in hypoacetylated regions (18, 51). H3K4me3 also correlated with transcriptional activity across the β-globin locus in cultured cells (19). We report that in skeletal muscle, where MHC gene expression undergoes alterations in expression during unloading, a similar pattern of histone acetylation is observed. The IIx and IIb MHC, which have very little transcriptional activity in the Sol, are relatively hypoacetylated. Upon induction of the IIx and IIb MHC genes in HS Sol, these genes become hyperacetylated. In the tissue comparison between the Pla and Sol, differential patterns of acetylation are also observed. The type I MHC is hyperacetylated in the Sol relative to the Pla, while the opposite is observed in the fast IIx and IIb MHCs, i.e., the IIx and IIb are hyperacetylated in the Pla relative to the Sol.
Given that the finding that relative enrichment of histone H3 acetylation at the type I, IIx, and IIb MHC genes corresponds with each MHC's respective expression level in each muscle type examined, it is surprising that in HS Sol, where IIa MHC mRNA is significantly decreased compared with Sol, we did not observe a corresponding deacetylation at the IIa MHC. This suggests that transcriptional downregulation of the IIa MHC is not dependent on histone deacetylation. Furthermore, enrichment levels of H3K4me3 were also not changed at the IIa MHC. This seeming contradiction may indicate an alternative regulatory mechanism for the IIa MHC in unloaded slow muscle. Indeed, as previously reported, antisense RNA is transcribed across the IIa MHC and its promoter at significantly increased levels in the inactive Sol (31). This antisense RNA likely mediates transcriptional silencing of the IIa MHC (31). Thus, the effect of the antisense RNA may negate the need for a histone acetylation/deacetylation strategy to regulate the IIa MHC in the unloaded slow myofiber. However, it is still not clear by what mechanism the antisense RNA exerts control. Preliminary ChIP analyses with histone modifications known to repress transcriptional activity, H3K9me2, H3K9me3, and H3K27me3 were not enriched at the IIa MHC gene in HS Sol vs. Sol (data not shown). Perhaps, other histone modifications or methylation of the DNA itself may mediate repression of IIa MHC induced by antisense RNA.
The pattern of H3K4 trimethylation across the three muscle types studied was similar to that of histone H3 acetylation for the IIx and IIb MHCs, and followed the same pattern as mRNA/pre-mRNA expression. In type I MHC there was a reduction in both mRNA and H3ac in HS Sol as compared with Sol, but H3K4me3 was not correspondingly decreased. While type I MHC pre-mRNA and mRNA were downregulated, the gene is apparently not fully repressed, because transcripts continue to accumulate, indicating that transcription is still active and merely attenuated in HS Sol (see Fig. 1). This suggests that H3K4me3 at the type I MHC is less sensitive to the attenuated transcription than H3ac. To address this, we examined H3K4me3 (and H3ac) under conditions where transcription of the type I MHC is almost completely abolished. This model, where HS is combined with thyroid hormone (T3) treatment, resulted in a significant reduction in both H3K4me3 and H3ac at the type I MHC gene compared with Sol. This suggests that the lack of differential enrichment of H3K4me3 in Sol and HS Sol is due to the maintenance of transcription initiation of the type I MHC gene in HS Sol, albeit with reduced transcriptional activity (i.e., pre-mRNA). This is consistent with previously defined roles for H3K4me3 in transcription initiation (13, 30).
H3K4me3 along with K36 methylation is implicated in a regulatory phase before transcription elongation to ensure controlled onset of transcription (30). This is supported by a whole genome ChIP-chip approach in human cells, where H3K4me3 is shown to be associated with transcription initiation, but not elongation (13). Furthermore, a H3K4me3-specific effector, the bromodomain PHD finger transcription factor (BPTF), is a subunit of nucleosome remodeling factor (NURF), a chromatin remodeling complex which facilitates transcription (50). It is proposed that H3K4me3 serves as a docking site for NURF, recruited to promoters by sequence-specific transcription factors, which mediates transcription initiation via energy-dependent disruption of nucleosomes (50). Given this role for H3K4me3, it may not be surprising that this histone modification is not diminished in the type I MHC of HS Sol, since transcription initiation is still necessary. Conversely, expression is strongly increased in the IIx and IIb MHCs in HS Sol, compared with Sol, and H3K4me3 is proportionally increased, perhaps resulting in more frequent transcription initiation at each gene and/or indicating that a greater number of myonuclei are actively transcribing IIx and IIb MHC.
H3K4me3 has been shown to be positively correlated with H3 acetylation using ChIP methods (13, 38, 48) and mass spectroscopy (17, 45, 52). H3K4me3 and H3ac can also exist on the same H3 tail (15, 17, 45). Hazzalin and Mahadevan (15), in a series of experiments, show that all detectable H3K4me3 is targeted for rapid turnover of acetylation at H3. Thus, H3K4me3 may serve as a target for continuous acetylation and deacetylation by HATs and HDACs (15). All of the MHC genes in all of the muscle tissues examined had some degree of both H3ac and H3K4me3. In response to 7 days of HS, the slow soleus muscle is phenotypically remodeled, and we observed induction of the IIx and IIb MHC genes. This corresponds positively with both increased histone H3 acetylation and trimethylation at H3K4. Upon repression of the type I MHC with HS, histone H3 is deacetylated in HS Sol. These findings are consistent with the model of rapid turnover of acetyl groups at sites that are also trimethylated at H3K4. Rapid turnover of acetyl groups may have important implications for genes with regard to their response to treatment with HDAC inhibitors.
Transcriptional therapies using HDAC inhibitors to influence skeletal muscle remodeling or to treat muscular dystrophies are being actively explored (4, 20, 29, 33). However, there are functional implications to histones subject to dynamic turnover of acetylation, as opposed to more stably enhanced acetylation, which occurs when HDACs are inhibited (8, 9). Hazzalin and Mahadevan (15) have demonstrated that the opposing actions of HATs and HDACs are necessary for normal expression of some genes. For example, upon treatment of mouse fibroblasts with the HDAC inhibitor trichostatin A, which allows the activity of HATs to be unopposed, the induced expression of c-jun was inhibited, though the histones of c-jun remained hyperacetylated (15). These results illustrate the complex manner by which inhibitors of HDACs can influence gene expression, by resulting in either activation or repression, depending on the target gene. As research progresses in this area, it will be important to consider dynamic turnover of acetyl groups in MHC chromatin and the potential unintended consequences of unopposed acetylation at MHC loci that may require dynamic turnover of acetylation for proper regulation.
The fact the H3K4me3 and H3ac can occur on the same histone tail, and are associated with active transcription, supports the emerging view of a histone code defined by multivalent histone modifications. Multivalency may impart greater specificity to the coactivators that recognize specific histone modifications via bromodomains, which recognize acetylated lysine, and PHD finger domains, which recognize H3K4me3 (35). Importantly, many chromatin-associated proteins have been identified that contain both bromodomains and PHD fingers (34). For example, one identified effector, BPTF, contains the domains to recognize both histone marks (24). Furthermore, multivalent binding may result in enhanced affinity of such interactions, while remaining more dynamic than a correspondingly tight monovalent interaction (35).
The role of histone methylation on gene transcription has, until recently, centered on that of a highly stable histone modification, with its primary role in the storage of epigenetic information, and propagation of such information across cell divisions as well as in epigenetic inheritance (1, 39, 47). Thus, histone lysine methylation was thought to be primarily a one-way process carried out by histone methyltransferases. However, histone methylation is now known to be enzymatically reversible and dynamically regulated (39). For example, a mechanistic role for dynamic regulation of H3K4me3 has been described for gene induction in yeast (30). Little is known about the regulation of demethylation, as only recently has the first histone demethylase been identified and characterized (40). More recently yet, a new family of lysine demethylases, the Jumonji family, has been identified that can demethylate trimethylated histone tails, including a subfamily of these demethylases that can specifically revert H3K4me3 (1, 39). This has shifted the thinking of histone trimethylation to provide the basis for a dynamic view of H3K4me3 in the regulation of gene expression. Our data on the dynamically regulated MHC genes would support this view. We report that with strong upregulation of the IIx and IIb MHCs in HS Sol as compared with Sol, H3K4me3 was also strongly increased at the 5′-end of these genes. We also observed a loss of H3K4me3 in the type I MHC under conditions (HS + T3), where the activity of this gene is markedly repressed. Clearly, regulation of H3K4me3 does occur in correspondence with expression of the MHC genes.
In conclusion, we provide evidence in this report that the chromatin immunoprecipitation assay can be used to accurately examine in vivo native histone modifications at specific genomic sites in skeletal muscle. We show that H3 acetylation and H3K4me3 are dynamically altered in conjunction with slow to fast shifts in MHC gene expression. Furthermore, the maintenance of the slow versus fast MHC phenotype is linked to the MHC chromatin environment by the degree of H3 acetylation and H3K4me3, which corresponds to MHC transcription levels in different fiber types. In addition to the increase in histone acetylation-induced accessibility of DNA by transcription factors, the recruitment of effectors to marks such as H3K4me3 may impart MHC-specific activation in response to specific loading and/or hormonal states. Thus, epigenetic mechanisms are another factor to be considered in the coordinated regulation of the types I, IIa, IIx, and IIb MHC genes. This initial survey of H3 acetylation and H3K4me3 in the MHC gene locus provides a starting point for further research into the factors that influence MHC gene regulation in their natural in vivo setting. Skeletal muscle is particularly sensitive to many stimuli, including loading state, that provide constant feedback on gene expression and cell signaling programs. Therefore, the sum influence of the entire system can only be faithfully represented in intact muscle. For this reason, the ChIP assay should prove to be an extremely useful tool to examine protein-DNA interactions under varying environmental conditions in skeletal myofibers.
GRANTS
This research was supported by National Institutes of Health Grants AR-30346 (to K. M. Baldwin) and T32-AR-047752.
Acknowledgments
The authors thank Jiang H. Weihua, LiYing Zhang, and Anqi X. Qin for excellent technical assistance; and Alvin M. Yu, Phuc D. Tran, Daniel Jimenez, Sandy Liu, Tiffany Yu, Marinelle Camilon, Bryce Buchowicz, Kareem Barghouti, and Allen Chao for exceptional animal care and tissue collection.
REFERENCES
- 1.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev 18: 159–168, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA 51: 786–794, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold HH, Winter B. Muscle differentiation: more complexity to the network of myogenic regulators. Curr Opin Genet Dev 8: 539–544, 1998. [DOI] [PubMed] [Google Scholar]
- 4.Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P, Cizman Z, Di Prospero NA, Pellizzoni L, Fischbeck KH, Sumner CJ. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest 117: 659–671, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin KM, Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol 90: 345–357, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14: 167–196, 1998. [DOI] [PubMed] [Google Scholar]
- 8.Bresnick EH, John S, Berard DS, LeFebvre P, Hager GL. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci USA 87: 3977–3981, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 23: 289–296, 2006. [DOI] [PubMed] [Google Scholar]
- 10.De la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays 27: 164–175, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Di Maso NA, Haddad F, Zeng M, McCue SA, Baldwin KM. Role of denervation in modulating IIb MHC gene expression in response to T3 plus unloading state. J Appl Physiol 88: 682–689, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Elferink CJ, Reiners JJ Jr. Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477, 1996. [DOI] [PubMed] [Google Scholar]
- 13.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddad F, Qin AX, Giger JM, Guo H, Baldwin KM. Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR. BMC Biotechnol 7: 21, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol 3: e393, 2005. [DOI] [PMC free article] [PubMed]
- 16.Huey KA, Roy RR, Baldwin KM, Edgerton VR. Temporal effects of inactivity on myosin heavy chain gene expression in rat slow muscle. Muscle Nerve 24: 517–526, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry: LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J Biol Chem 282: 27923–27934, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Kim A, Dean A. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proc Natl Acad Sci USA 101: 7028–7033, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol Cell Biol 27: 1271–1279, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS, Fielitz J, McAnally J, Shelton JM, Lemon DD, McKinsey TA, Richardson JA, Bassel-Duby R, Olson EN. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol 28: 3600–3609, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation 113: 2579–2588, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell 117: 721–733, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar G, Johnson IM, Kale S, Raghow R. Epigenetic regulation of cardiac muscle-specific genes in H9c2 cells by interleukin-18 and histone deacetylase inhibitor m-carboxycinnamic acid bis-hydroxamide. Mol Cell Biochem 312: 47–60, 2008. [DOI] [PubMed] [Google Scholar]
- 26.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev 11: 497–504, 2001. [DOI] [PubMed] [Google Scholar]
- 27.Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci 8: 313–321, 2005. [DOI] [PubMed] [Google Scholar]
- 28.Mellor J, Dudek P, Clynes D. A glimpse into the epigenetic landscape of gene regulation. Curr Opin Genet Dev 18: 116–122, 2008. [DOI] [PubMed] [Google Scholar]
- 29.Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V, Fortuni S, Straino S, Sampaolesi M, Di Padova M, Illi B, Gallinari P, Steinkuhler C, Capogrossi MC, Sartorelli V, Bottinelli R, Gaetano C, Puri PL. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med 12: 1147–1150, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18: 723–734, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Pandorf CE, Haddad F, Roy RR, Qin AX, Edgerton VR, Baldwin KM. Dynamics of myosin heavy chain gene regulation in slow skeletal muscle: role of natural antisense RNA. J Biol Chem 281: 38330–38342, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest 117: 2459–2467, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411, 2002. [DOI] [PubMed] [Google Scholar]
- 37.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev 15: 528–535, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25: 1–14, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953, 2004. [DOI] [PubMed] [Google Scholar]
- 41.Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol 41: 185–198, 2009. [DOI] [PubMed] [Google Scholar]
- 42.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 403: 41–45, 2000. [DOI] [PubMed] [Google Scholar]
- 43.Tang H, Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc Natl Acad Sci USA 103: 16977–16982, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Macpherson P, Marvin M, Meadows E, Klein WH, Yang XJ, Goldman D. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol Biol Cell 20: 1120–1131, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci USA 104: 2086–2091, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomason DB, Herrick RE, Surdyka D, Baldwin KM. Time course of soleus muscle myosin expression during hindlimb suspension and recovery. J Appl Physiol 63: 130–137, 1987. [DOI] [PubMed] [Google Scholar]
- 47.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell 125: 213–217, 2006. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright C, Haddad F, Qin AX, Baldwin KM. Analysis of myosin heavy chain mRNA expression by RT-PCR. J Appl Physiol 83: 1389–1396, 1997. [DOI] [PubMed] [Google Scholar]
- 50.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90, 2006. [DOI] [PubMed] [Google Scholar]
- 51.Yin W, Barkess G, Fang X, Xiang P, Cao H, Stamatoyannopoulos G, Li Q. Histone acetylation at the human beta-globin locus changes with developmental age. Blood 110: 4101–4107, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K, Siino JS, Jones PR, Yau PM, Bradbury EM. A mass spectrometric “Western blot” to evaluate the correlations between histone methylation and histone acetylation. Proteomics 4: 3765–3775, 2004. [DOI] [PubMed] [Google Scholar]