skeletal muscle is a highly plastic tissue, capable of responding and adapting to a variety of physiological stimuli, e.g., muscle loading and unloading. Multiple functional phenotypes of skeletal muscle exist throughout the body at the same time. For example, muscles required for antigravity support of the skeleton or sustained locomotion are generally slow-contracting, low force-generating, and relatively resistant to fatigue, while other muscles required for quick, powerful movements are generally fast-contracting, high force-generating, and easily fatigued. In fact, skeletal muscle phenotypes have been characterized using combinations of muscle properties, including metabolic, fatigability, color, innervation, or predominant contractile protein expression. Myosin heavy chain (MHC) is the most abundant protein in skeletal muscle, comprising ∼25% of the total protein content, and is a major component of the complex responsible for generating contractile force in skeletal muscle. At least nine MHC isoforms, each transcribed from their own gene, exist in mammalian striated muscle; but only slow type I MHC and various isoforms of fast type II (IIa, IIx/d, and IIb MHC isoforms) are present in adult limb skeletal muscle that is not undergoing regeneration (16). It is well established that skeletal muscle phenotype, particularly MHC gene expression, can be altered by different states of muscular activity or inactivity, as well as various hormonal and metabolic factors (1).
In response to altered muscular activity, skeletal muscle undergoes MHC remodeling to adapt to the new physiological demands on the tissue. The final composition of various MHC protein isoforms after adaptation is dependent on the species, on the environmental stimulus type and intensity (whether it is physiological or supra-physiological), and on contractile activity (whether it is increased or decreased). Resistance training switches rat MHC from type IIb to IIx MHC in red and white portions of rat gastrocnemius muscle (4). Importantly, no increase in type I MHC occurs in response to physiological resistance training. However, supra-physiological models of muscle hypertrophy (functional overload) do increase type I fiber percentage (17) in rats. Similarly, endurance training, a physiological stimulus, for <3 mo exhibits no increase in type I MHC (13; see discussion in Ref. 9), whereas, in contrast, supra-endurance models (chronic stimulation for >12 h/day) have major increases in the percentage of type I MHC (13). Physiological decreases in muscle loading by hindlimb suspension or spaceflight do decrease type I fiber percentage in rodents (3, 5) and humans (8, 18, 19). Taken together, the question then is how does each nucleus decide which MHC gene to express and not to express in a muscle fiber with multiple nuclei during reductions in contractile activity?
Six of the nine MHC genes are arranged in tandem on the same chromosome, and their order and orientation are highly conserved. Multiple control mechanisms have been shown to exist beyond transcription factors. Surprisingly, while pre-mRNA (10), alternative splicing (7), antisense RNA (11), and intergenic transcription (15) are all known to regulate MHC, little is known about whether epigenetic mechanisms also regulate MHC gene expression in skeletal muscle with altered muscular activity. Pandorf et al. (12) describe a novel mechanism by which epigenetic modifications play a role in the expression of MHC genes in response to altered muscular activity (12).
Until recently, chromatin has rarely been thought of as much more than just genomic DNA and protein packaging. However, an emerging field in biological research over the past decade, termed “epigenetics,” has given new-found respect to chromatin structure as being a major player in the control of transcriptional activity and gene expression. Epigenetic modifications, unlike genetic mutations or single nucleotide polymorphisms, do not result in changes in the DNA sequence. Instead, DNA structure is indirectly modified by histone modifications (e.g., acetylation or methylation) or directly modified by methylation of the cytosine base in cytosine-guanine dinucleotides (CpG) of DNA to alter the expression of genes. CpG methylation of DNA is a stable covalent modification more associated with disease states, such as cancer or atherosclerosis (2, 14), while histone modifications are more akin to protein phosphorylation. Histone modifications can be both transient, rapidly responding to changes within the cell or surrounding environment, or stably maintained throughout life when obtained in early or fetal life (6). Histone acetylation can occur at many different lysine residues (e.g., lysine 4, 9, or 27), creating a negative electrostatic charge that repels the negatively charged DNA backbone, thus producing a loosely packed chromatin that allows easier access for transcription factors and the RNA polymerase complex to increase transcription (Fig. 1), also providing a site for recruitment of other proteins to the promoter region of a given gene. Conversely, histone deacteylation results in a compact, tightly bound histone:DNA complex and general repression of gene transcription (6).
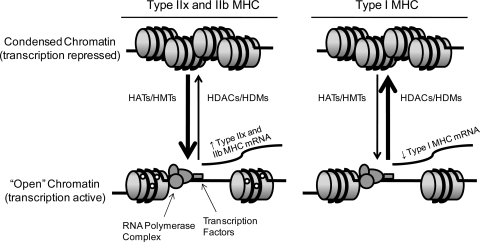
Fig. 1.
Schematic representing epigenetic histone modifications occurring in the slow fiber-type soleus muscle in response to reduced muscular activity/unloading (hindlimb suspension). Upon induction of hindlimb suspension, histone acetylation and/or histone methylation (left), by histone acetyltransferases (HATs) and histone methyltransferases (HMTs), respectively, are associated with an “open” chromatin structure and increased transcriptional activity of type IIx and IIb myosin heavy chain (MHC) genes. On the other hand, histone deacetylation and/or histone demethylation (right), by histone deacetylases (HDACs) and histone demethylases (HDMs), respectively, are associated with a condensed chromatin structure and repressed transcriptional activity of the type I MHC gene in response to hindlimb suspension. ○, Acetyl or methyl groups covalently bound to histones.
The acetylation status of core histones is based on the balance of histone acetyltransferase (HAT) and histone deacetylase (HDAC) activity. Although it has been known that skeletal muscle contraction during exercise can regulate the activity of HATs and HDACs, this report by Pandorf et al. (12) is the first to show with direct evidence that histone modifications occur in skeletal muscle at the locus of MHC genes, concomitantly with changes in MHC gene expression. Using chromatin immunoprecipitation (ChIP), the authors demonstrate that both histone H3 acetylation and histone H3 methylation are directly related to the steady-state transcriptional activity of the MHC genes in the fast fiber-type plantaris and in the slow fiber-type soleus muscles under normal conditions. Of particular interest is the finding that the acetylation and methylation status at specific sites of histone H3 also directly coincides with the altered transcriptional activity of type I, IIx, and IIb MHC genes in the soleus in response to reduced muscular activity/unloading (hindlimb suspension).
These findings have significant physiological relevance in skeletal muscle biology because they extend our current understanding of the molecular mechanisms that regulate skeletal muscle remodeling and MHC gene expression with altered muscular activity. Also, Pandorf et al. (12) demonstrate the feasibility of using ChIP assays to analyze native chromatin structure in frozen samples as it pertains to transcriptional activity of skeletal muscle genes. Certainly, studies elucidating the mechanisms by which specific HATs and HDACs change their activity in response to a physiological stimulus, such as hindlimb unloading or endurance exercise, are needed. Additionally, answering broad questions such as whether epigenetics play a role in the regulation of other genes involved in skeletal muscle remodeling, and further yet, whether epigenetic regulation is involved in muscle phenotypes with pathological consequences, such as the failed regrowth of muscle from atrophy with aging, muscular dystrophies, or cardiac myopathies, is beginning to come within reach thanks to studies such as this one.
GRANTS
This work was supported by National Institutes of Health Grant AG-18780 (to F. W. Booth), American College of Sports Medicine (to M. J. Laye), and American Heart Association (to M. J. Laye).
REFERENCES
- 1.Baldwin KM, Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol 90: 345–357, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2, Suppl 1: S4–S11, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Caiozzo VJ, Baker MJ, McCue SA, Baldwin KM. Single-fiber and whole muscle analyses of MHC isoform plasticity: interaction between T3 and unloading. Am J Physiol Cell Physiol 273: C944–C952, 1997. [DOI] [PubMed] [Google Scholar]
- 4.Caiozzo VJ, Haddad F, Baker MJ, Baldwin KM. Influence of mechanical loading on myosin heavy-chain protein and mRNA isoform expression. J Appl Physiol 80: 1503–1512, 1996. [DOI] [PubMed] [Google Scholar]
- 5.Caiozzo VJ, Haddad F, Baker MJ, Herrick RE, Prietto N, Baldwin KM. Microgravity-induced transformations of myosin isoforms and contractile properties of skeletal muscle. J Appl Physiol 81: 123–132, 1996. [DOI] [PubMed] [Google Scholar]
- 6.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 23: 289–296, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Dennehey BK, Leinwand LA, Krauter KS. Diversity in transcriptional start site selection and alternative splicing affects the 5′-UTR of mouse striated muscle myosin transcripts. J Muscle Res Cell Motil 27: 559–575, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Edgerton VR, Zhou MY, Ohira Y, Klitgaard H, Jiang B, Bell G, Harris B, Saltin B, Gollnick PD, Roy RR. Human fiber size and enzymatic properties after 5 and 11 days of spaceflight. J Appl Physiol 78: 1733–1739, 1995. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Roves PM, Huss J, Holloszy JO. Role of calcineurin in exercise-induced mitochondrial biogenesis. Am J Physiol Endocrinol Metab 290: E1172–E1179, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. II. Molecular markers of protein deficits. J Appl Physiol 95: 791–802, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Pandorf CE, Haddad F, Roy RR, Qin AX, Edgerton VR, Baldwin KM. Dynamics of myosin heavy chain gene regulation in slow skeletal muscle: role of natural antisense RNA. J Biol Chem 281: 38330–38342, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Pandorf CE, Haddad F, Wright C, Bodell PW, Baldwin KM. Differential epigenetic modifications of histones at the myosin heavy chain genes in fast and slow skeletal muscle fibers and in response to muscle unloading. Am J Physiol Cell Physiol (April 15, 2009). doi: 10.1152/ajpcell.00075.2009. [DOI] [PMC free article] [PubMed]
- 13.Pette D Historical perspectives: plasticity of mammalian skeletal muscle. J Appl Physiol 90: 1119–1124, 2001. [DOI] [PubMed] [Google Scholar]
- 14.Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res 43: 985–991, 1999. [DOI] [PubMed] [Google Scholar]
- 15.Rinaldi C, Haddad F, Bodell PW, Qin AX, Jiang W, Baldwin KM. Intergenic bidirectional promoter and cooperative regulation of the IIx and IIb MHC genes in fast skeletal muscle. Am J Physiol Regul Integr Comp Physiol 295: R208–R218, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76: 371–423, 1996. [DOI] [PubMed] [Google Scholar]
- 17.Stone J, Brannon T, Haddad F, Qin A, Baldwin KM. Adaptive responses of hypertrophying skeletal muscle to endurance training. J Appl Physiol 81: 665–672, 1996. [DOI] [PubMed] [Google Scholar]
- 18.Trappe S, Costill D, Gallagher P, Creer A, Peters JR, Evans H, Riley DA, Fitts RH. Exercise in space: human skeletal muscle after 6 months aboard the International Space Station. J Appl Physiol 106: 1159–1168, 2009. [DOI] [PubMed] [Google Scholar]
- 19.Zhou MY, Klitgaard H, Saltin B, Roy RR, Edgerton VR, Gollnick PD. Myosin heavy chain isoforms of human muscle after short-term spaceflight. J Appl Physiol 78: 1740–1744, 1995. [DOI] [PubMed] [Google Scholar]