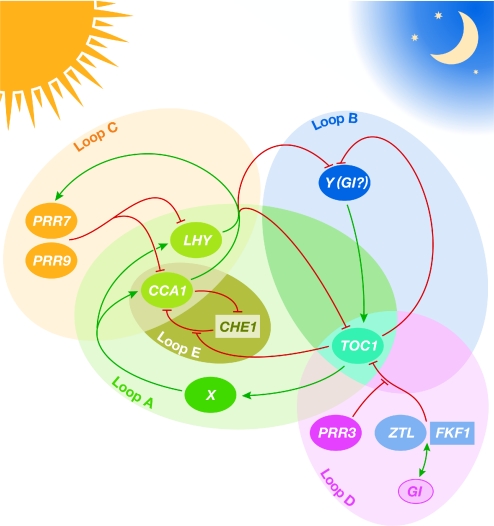
Figure 2.
Model of the Arabidopsis circadian clock. The circadian oscillator consists of a series of interlocking feedback loops (A–E). Genes newly reported to function in the clock are shown within rectangles. Loop A represents the transcriptional feedback loop that was identified initially and contains the morning-phased transcription factors CCA1 and LHY, which negatively regulate TOC1. Component X, the existence of which has been inferred from mathematical modelling, induces the transcription of CCA1 and LHY. CHE1 could provide some of the functionality represented by X. Two evening-phased genes, TOC1 and the yet-unidentified component Y, make up loop B. Morning-phased PRR7, PRR9, CCA1 and LHY make up loop C. As reported at the meeting, FKF1 works with ZTL to regulate TOC1 negatively, and this process is, in turn, regulated by GI and PRR3, thereby constituting loop D. The existence of loop E, which provides a link between TOC1 and CCA1, was proposed for the first time at the meeting. Some genes implicated in clock function have been omitted for clarity. Adapted, with permission, from Harmer (2009). CCA1, CIRCADIAN CLOCK-ASSOCIATED 1; CHE1, CCA1 HIKING EXPEDITION 1; FKF1, FLAVIN-BINDING KELCH REPEAT, F-BOX 1; GI, GIGANTEA; LHY, LATE ELONGATED HYPOCOTYL; PRR, PSEUDORESPONSE REGULATOR; TOC1, TIMING OF CAB EXPRESSION 1; ZTL, ZEITLUPE.