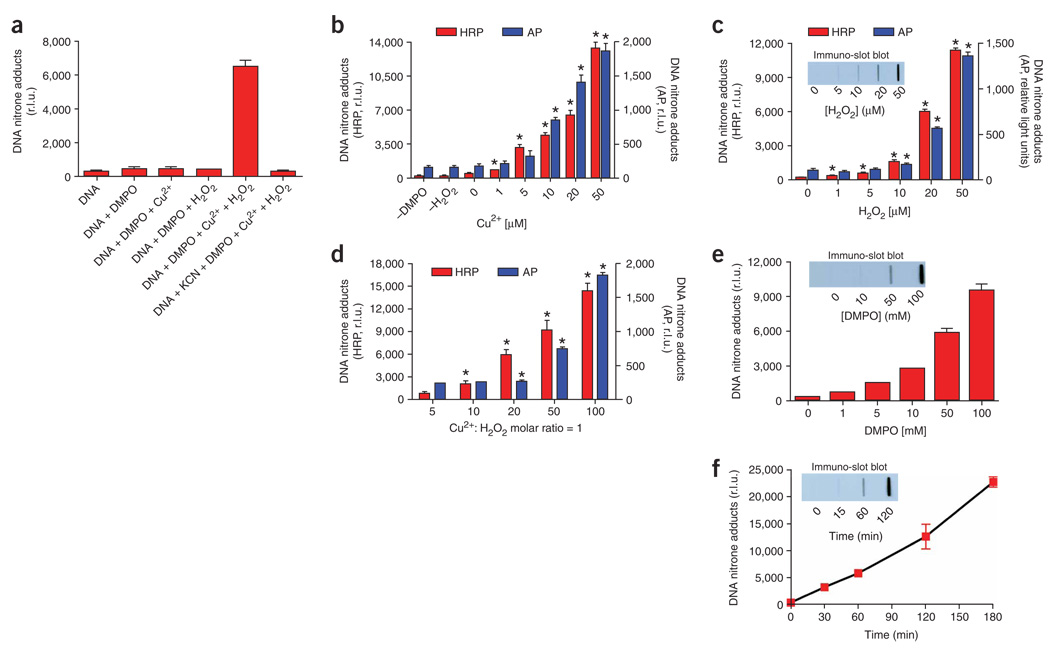
Figure 3.
Immuno-spin trapping analysis of DNA radicals. (a) We mixed the following components: 20 µM DNA (as nucleotides), 20 µM Cu2+ (as chloride salt), 20 µM H2O2 and/or 50 mM DMPO in 100 mM chelexed PB (pH 7.4), in a total volume of 300 µl. We added a 10-X stock DNA solution (30 µl) and the other components from a 100-X stock solution (3 µl). After 1 h incubation, we stopped the reaction with 3 µl of 1 M KCN or 100 mM DTPA. Stopping the reaction with either reagent gave similar results. After stopping, we froze the samples until analysis. (b) The procedure was as described for (a), but we added different concentrations of Cu2+ or omitted one of the components in the reaction mixture. *P < 0.05 with respect to “zero” Cu2+. (c) The procedure was as described for (b), but we varied the final concentration of H2O2 in the reaction mixtures. *P <0.05 (with respect to “zero” H2O2. (d) We added 20 µM DNA, 50 mM DMPO, and equal concentrations of Cu2+ and H2O2 (e.g., “5” corresponds to 5 µM Cu2+ and 5 µM H2O2, and so on). *P < 0.05 with respect to “5”(e.g., 5 µM Cu2+:5 µM H2O2). (e) The procedure was as described for (c), but we added different final concentrations of DMPO. (f) We mixed 20 µM DNA, 20 µM Cu2+, 50 mM DMPO and 20 µM H2O2, and then added 3 µl of 1 M KCN to stop the reaction at different times after the addition of H2O2. For time “zero”, we added KCN just before H2O2. We performed the immuno-spin trapping analysis as described in the PROCEDURE. Insets show the immuno-slot blot corresponding to the samples analyzed by ELISA in the main graph. We compared AP (blue bars) with our improved developing system using HRP (red bars). We developed the immuno-slot blot using the HRP system. Data show the mean ± s.e.m. from three separate experiments in quadruplicate (n = 12).