Figure 1.
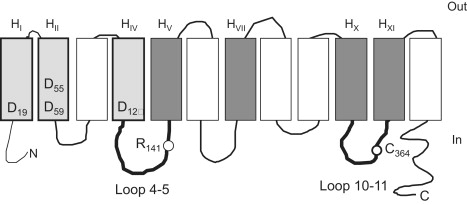
Predicted secondary structural model of MelB. The 12 successive transmembrane helices are represented by boxes and labeled using the H letter and a roman number subscript. Shaded boxes indicate helices putatively lining the cosubstrate binding sites (light gray, Na+ site; dark gray, sugar site). Amino acids of interest for this study are labeled using the single-letter amino acid code and a number subscript corresponding to their position on the primary MelB amino acid sequence. The aspartic acid residues distributed in HI, HII, and HIV are important for Na+ binding. Arg141 is located in loop 4-5. Cys364 in loop 10-11 is the NEM target. The model is from Pourcher et al. (9).
