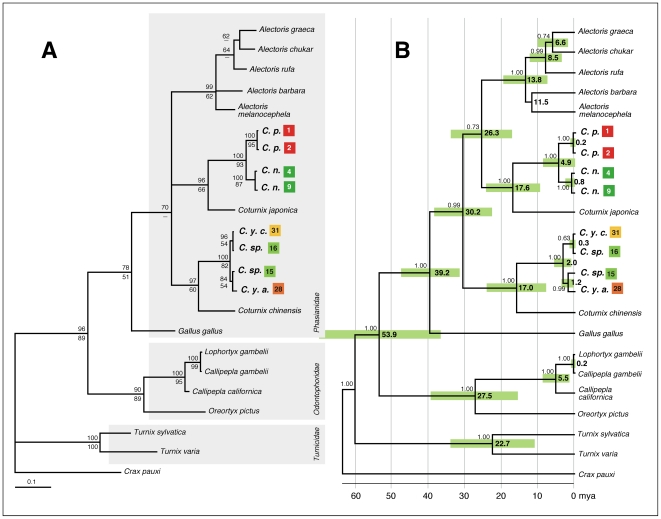
Figure 3. Phylogenetic relationships and split times between Coturnix sp.
A. A distance neighbour-joining tree was constructed with 456 bp of mitochondrial cytochrome b sequence in PAUP* v4.0beta10 using the TrN+I+G model of nucleotide substitution where the proportion of invariable (I) sites was calculated to be 0.438 and the Gamma distribution shape parameter (G) was 0.755. Node bootstrap values were determined using both Distance (above line) and ML (below line) -based methods. Only values greater than 50% are shown. Crax Pauxi from the Cracidae family was used as outgroup. B. BEAST v1.4.8 maximum clade credibility tree. Divergence times are given in millions of years ago (mya; 95% HPD indicated by the green boxes). Node posterior probability values were calculated in BEAST v1.4.8 and values greater than 0.6 are shown above the branch lines. Abbreviations, colours, and numbers are as outlined in Figure 2.