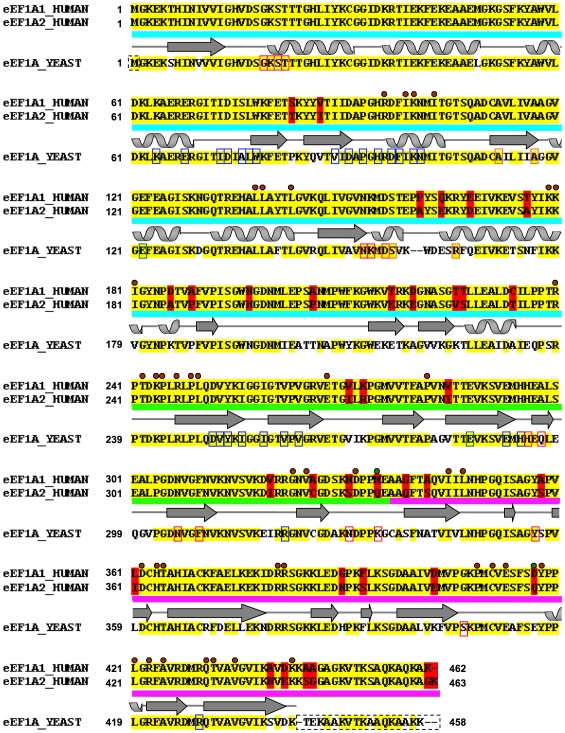
Figure 1. Sequence alignment between human eEF1A1 and eEF1A2 and yeast template.
The pair-wise sequence alignment between human eEF1A1 and eEF1A2 is shown: identical residues (yellow background), variant residues (red background). The aligned yeast eEF1A template is shown below with identical residues to the human sequences highlighted (yellow background) and any variant position between yeast and either human sequence shown with a white background. The two human sequences share 92% sequence identity with each other and each show ∼81% sequence identity with the yeast protein. The domain boundaries (domain I: cyan; domain II: green; domain III: pink), and STRIDE [44] secondary structure assignment is traced above the yeast template sequence (arrows = beta-strands; coils = alpha-helices). The amino acid residues involved in domain-domain contacts are indicated with a brown circle (green circle for non-identical equivalent residues between two human variants); those involved in the binding of C-terminal fragment eEF1Bα are indicated on the yeast sequence with blue rectangles; residues involved in GDP-binding indicated in pink rectangles; and those disordered in the yeast crystal structure are indicated with a dashed rectangle. Yeast mutagenesis data and motifs are highlighted on its sequence: mutations involved in actin bundling/disorganization (red rectangles) [70], [71]; mutations that affect translational fidelity (green rectangles) [77]; mutations that reduce dependence on eEF1B (orange rectangles) [78], [79].