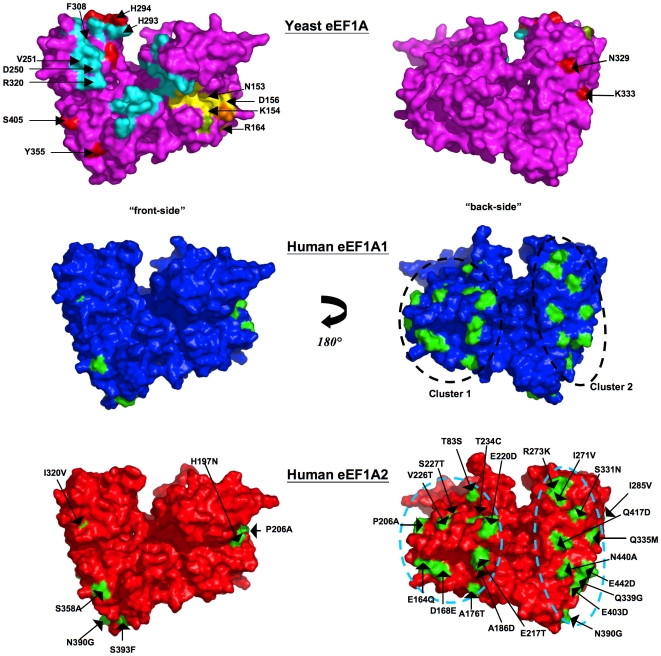
Figure 3. Location of variations in amino acids mapped onto surface.
Two equivalent views rotated by 180° about the y-axis depicting a surface rendition of the yeast eEF1A crystal structure colored magenta (top panel), and the 3-D models of eEF1A1 colored blue (middle panel) and eEF1A2 colored red (bottom panel). Locations of exposed variant side-chains are mapped onto the surface of the two model proteins (colored green) and labeled on the eEF1A2 model - the variant residue from eEF1A1 is shown on the right-hand side of the label. The two sub-clusters are apparent in this representation. The location of the C-terminal eEF1Bα-binding site (cyan) [38] and GDP-binding site (yellow) [39] is mapped on the crystal structure. Also highlighted (red) on its surface are: mutations that reduce actin disorganization induced by overexpression of eEF1A, inhibit actin-bundling without altering translation in vivo, and reduce actin-bundling [70], [71]. There are no variants in proximity to those residues implicated on the basis of mutagenesis to be involved in translational fidelity (green) [77]. However, two variant positions in humans - Gln164Glu and Glu168Asp are in close proximity to Arg166 – a conservative mutation for the equivalent residue in yeast (Arg164Lys) was shown to reduce dependence on eEF1B (orange) [78], [79]. Gln164Glu and Glu168Asp, however, both retain their main-chain to main-chain H-bonds with Arg166. Note: for clarity the three proposed aminoacyl-tRNA-binding residues [38] are not shown, since they overlap with already highlighted positions implicated in binding eEF1Bα (His293 and Arg320) and actin (His294).