Abstract
The aryl hydrocarbon receptor (AHR) is a protein best known for its role in mediating toxicity. Over 30 years of research has uncovered additional roles for the AHR in xenobiotic metabolism and normal vascular development. Activation of the AHR has long been known to cause immunotoxicity, including thymic involution. Recent data suggesting a role for the AHR in regulatory T-cell (Treg) and T-helper 17 (Th17) cell development have only added to the excitement about this biology. In this review, we will attempt to illustrate what is currently known about AHR biology in the hope that data from fields as diverse as evolutionary biology and pharmacology will help elucidate the mechanism by which AHR modifies immune responses. We also will discuss the complexities of AHR pharmacology and genetics that may influence future studies of AHR in the immune system.
Keywords: aryl hydrocarbon receptor, dioxin, immune function, thymus, T lymphocytes
Introduction
The aryl hydrocarbon receptor (AHR) is a protein of ancient origins. Phylogenetic analysis reveals that functional orthologues of the Ahr gene are present in living mammals, amphibians, reptiles and birds.1 For more than 30 years, the AHR has been studied as a receptor for environmental contaminants and as a mediator of chemical toxicity. Recently, an additional role for the AHR in normal vascular development has been identified. Longstanding literature on 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) toxicology, as well as a flurry of recent high-profile papers, has suggested a role for this protein in immunology. In this review, we will provide an overview of AHR signal transduction with an emphasis on providing information that may guide future studies aimed at delineating the role of this protein in human immunity and related disease.
The AHR signalling pathway
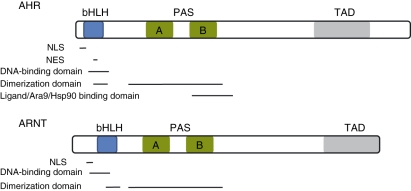
The AHR is a ligand-activated transcription factor from the Per-Arnt-Sim (PAS) superfamily of proteins.2 Analysis of the AHR reveals an N-terminal basic helix loop helix (bHLH) domain, a C-terminal variable domain, and a central PAS domain with two degenerate repeats (denoted repeat A and repeat B).3–5 The PAS domain of the AHR mediates heterodimerization with a structurally related protein known as the aryl hydrocarbon receptor nuclear translocator (ARNT), DNA recognition, ligand binding and chaperone interactions.5–7 Next to the PAS domain is a bHLH domain that is involved in DNA binding and support of dimerization.4,6,8 The C-terminal half is highly variable and responsible for differences in receptor molecular weight within and across species.2,5 The largely unstructured C-terminal region contains a transcriptionally active domain and potentially domains involved in receptor transformation2,5 (Fig. 1).
Figure 1.
Protein domains found in the aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT). The nuclear localization sequence (NLS) and nuclear export sequence (NES) are found within the basic helix loop helix (bHLH) region. The bHLH also plays a critical role in DNA binding. The characteristic domains in Per-ARNT-Sim (PAS) family members mediate heterodimerization and chaperone binding. The C-terminus is variable, but contains the transactivation domain (TAD) responsible for activating transcription after DNA binding.
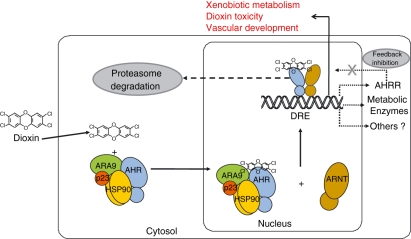
In the absence of bound agonist, the AHR is most commonly found in the cytoplasm in a complex with its chaperones heat shock protein 90 (Hsp90), P23 and aryl hydrocarbon receptor associated 9 (ARA9; aka AIP, XAP2).9,10,12 In addition to holding the receptor in a form able to bind ligand, Hsp90 prevents surreptitious nuclear translocation of the AHR.9 The cochaperone p23 stabilizes the AHR–Hsp90 interaction, while the ARA9 protein enhances AHR signalling by increasing the amount of properly folded AHR in the cytoplasm.10,12 Upon agonist binding, the AHR–chaperone complex translocates to the nucleus and binds the ARNT protein.13,14 The ARNT protein is structurally similar to the AHR (Fig. 1). This heterodimeric pairing yields a competent transcription factor within the nuclear compartment of cells (Fig. 2).
Figure 2.
The aryl hydrocarbon receptor (AHR) signalling pathway. Hydrophobic ligands enter the cell via diffusion through the cell membrane. Ligands bind to the AHR in the cytosol. Ligand binding causes conformational changes leading to nuclear localization sequence (NLS) exposure and the AHR complex translocates to the nucleus. In the nucleus, the AHR binds its heterodimeric partner, the aryl hydrocarbon receptor nuclear translocator (ARNT) and directs transcription from dioxin response elements (DREs), upstream of target genes. Signalling through the AHR is down-regulated by two means, the proteasome and a feed-back pathway involving the aryl hydrocarbon receptor repressor (AHRR). The AHRR is an AHR gene target and its expression is up-regulated by AHR signalling. Signalling by the AHR leads to three biological pathways referred to as the adaptive metabolic pathway, the toxic pathway, and the developmental pathway. ARA9, aryl hydrocarbon receptor associated 9; Hsp90, heat shock protein 90.
The AHR:ARNT heterodimer directs transcription of genes from dioxin-responsive enhancer elements (DREs) within the genome. These classic enhancers are found near AHR transcriptional targets. These primary targets are commonly known as the ‘AHR gene battery’. Regulation of this gene battery has been shown to be dependent upon a number of common coactivators. For example, histone acetyl transferases are recruited to the DRE-regulated promoters through coactivators such as nuclear receptor coactivator (SRC) and p300.15–17 Additionally, transcriptional cofactors, such as transacting transcription factor 1 (Sp1), receptor interacting protein 140 (RIP140), and nuclear receptor subfamily 0 group B member 2 (SHP), activate the transcriptional response of the AHR:ARNT heterodimer.18,19 Although the list of members of the AHR gene battery is still expanding, it includes those that encode xenobiotic metabolizing enzymes, such as cytochromes P450 (CYP) 1A1, CYP1A2, CYP1B1 and UDP glucuronosyltransferase 1 family polypeptide A6 (UGT1A6)20 (Table 1). Collectively these enzymes are well known for their important roles in the clearance of foreign chemicals.
Table 1.
Aryl hydrocarbon receptor (AHR)-regulated genes. The genes listed here were identified as differentially expressed upon 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. All samples were compared to vehicle-treated control samples. The official gene symbol, gene name and GenBank accession number are provided. To search for dioxin responsive enhancer (DRE) sequences, we pulled the upstream sequence for each gene from the University of California–Santa Cruz (UCSC) genome browser and searched for DRE sequences on either the + or − strand using MotifViz.112,113 The number of times the DRE consensus sequence, TNGCGTG, was identified within 15 000 bases upstream of the gene target is indicated in the DRE column. In the Gene expression column, ↑ indicates that the gene was upregulate by TCDD treatment. A ↓ indicates that the expression of the gene was downregulated by TCDD. The number in the Gene expression column indicates the fold change between the TCDD and vehicle control treated samples. The Cell type column indicates the cell population used in each published experiment. Double negative-recent thymic emigrants (DN-RTE), triple negative X T-cell population (TNX), graft versus host disease (GVHD).
| Gene symbol | Gene name | Genebank accession | DRE | Gene expression | Cell type | Reference |
|---|---|---|---|---|---|---|
| Cyp1a1 | CYP450, family 1, subfamily a, polypeptide 1 | NM_001136059.1 | 5 | 38↑ | Liver | 114 |
| Fabp5 | Fatty acid binding protein 5 | NM_010634.2 | 0 | 4↑ | Liver | 114 |
| Gsta2 | Glutathione S-transferase α2 | NM_008182.3 | 3 | 7↑ | Liver | 114 |
| Itgb1 | Integrin β 1 | NM_010578.1 | 0 | 3↑ | Liver | 114 |
| Notch1 | Notch homolog 1 | NM_008714.2 | 4 | 3↑ | Liver | 114 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | NM_008706.5 | 4 | 5↑ | Liver | 114 |
| Ugdh | UDP-glucose dehydrogenase | NM_009466.2 | 2 | 3↑ | Liver | 114 |
| Car3 | Carbonic anhydrase 3 | NM_007606.3 | 1 | 4↓ | Liver | 115 |
| Cdca5 | Cell division cycle associated 5 | NM_026410.3 | 3 | 9↑ | Liver | 115 |
| Cfdp1 | Craniofacial development protein 1 | NM_011801.1 | 3 | 4↑ | Liver | 115 |
| Epyc | Epiphycan | NM_007884.2 | 1 | 6↑ | Liver | 115 |
| Gadd45b | Growth arrest & DNA damage-inducible 45β | NM_008655.1 | 2 | 5↑ | Liver | 115 |
| Lpin2 | Lipin2 | NM_022882.3 | 2 | 3↑ | Liver | 115 |
| Mrpl37 | Mitochondrial ribosomal protein L37 | NM_025500.1 | 3 | 8↑ | Liver | 115 |
| Myc | Myelocytomatosis oncogene | NM_010849.4 | 2 | 4↑ | Liver | 115 |
| Pabpc2 | Poly(A) binding protein, cytoplasmic 2 | NM_011033.2 | 0 | 7↑ | Liver | 115 |
| Pak1ip1 | PAK1 interacting protein 1 | NM_026550.2 | 1 | 4↑ | Liver | 115 |
| Tiparp | TCDD-inducible poly(ADP-ribose) polymerase | NM_178892.5 | 0 | 10↑ | Liver | 115 |
| Tnfaip2 | Tumor necrosis factor, α-induced protein 2 | NM_009396.2 | 4 | 6↑ | Liver | 115 |
| Ahrr | AHR repressor | NM_009644.2 | 2 | 21↑ | CD4+ cells (GVHD) | 72 |
| Ccr4 | Chemokine (C-C motif) recptor 4 | NM_009916.2 | 4 | 5↑ | CD4+ cells (GVHD) | 72 |
| Ccr5 | Chemokine (C-C motif) recptor 5 | NM_009917.5 | 0 | 3↑ | CD4+ cells (GVHD) | 72 |
| Ccr9 | Chemokine (C-C motif) recptor 9 | NM_009913.5 | 3 | 5↑ | CD4+ cells (GVHD) | 72 |
| Gzmb | Granzyme B | NM_013542.2 | 1 | 6↑ | CD4+ cells (GVHD) | 72 |
| IL12rb2 | Interleukin 12 receptor, β2 | NM_008354.3 | 1 | 10↑ | CD4+ cells (GVHD) | 72 |
| Prdm1 | PR domain containing 1 | NM_007548.3 | 2 | 3↑ | CD4+ cells (GVHD) | 72 |
| Stat4 | Signal transducer & activator of transcription 4 | NM_011487.4 | 2 | 3↑ | CD4+ cells (GVHD) | 72 |
| Tgfb3 | Transforming growth factor β3 | NM_009368.2 | 0 | 13↑ | CD4+ cells (GVHD) | 72 |
| Acpp | Acid phosphatase, prostate | NM_207668.2 | 4 | 16↑ | TNX | 66 |
| Bcl9 | B-cell lymphoma 9 | NM_029933.3 | 3 | 20↑ | TNX | 66 |
| Ctxn1 | Cortexin 1 | NM_183315.2 | 2 | 12↑ | TNX | 66 |
| Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | NM_010501.2 | 3 | 10↑ | TNX | 66 |
| IL12rb1 | Interleukin 12 receptor β1 | NM_008353.2 | 1 | 13↑ | TNX | 66 |
| Itgb7 | Integrin β7 | NM_013566.2 | 2 | 9↑ | TNX | 66 |
| Klf2 | Kruppel like factor 2 | NM_008452.2 | 0 | 13↑ | TNX | 66 |
| Lgals3 | Lectin, galactoside-binding soluble 3 | NM_010705.2 | 3 | 55↑ | TNX | 66 |
| Olfr1265 | Olfactory receptor 1265 | NM_146343.1 | 0 | 8↑ | TNX | 66 |
| Ret | Ret proto-oncogene | NM_001080780.1 | 2 | 11↑ | TNX | 66 |
| Ssxb2 | Synovial sarcoma, X member B, breakpoint 2 | NM_001001450.4 | 0 | 12↑ | TNX | 66 |
| Tm9Sf4 | Transmembrane 9 superfamily protein member 4 | NM_133847.3 | 4 | 14↑ | TNX | 66 |
| Traf5 | TNF receptor-associated factor 5 | NM_011633.1 | 0 | 10↑ | TNX | 66 |
| Vps25 | Vacuolar protein sorting 25 | NM_026776.3 | 3 | 12↑ | TNX | 66 |
| Zcchc2 | Zinc finger, CCHC domain containing 2 | NM_001122675.1 | 1 | 9↑ | TNX | 66 |
| Cyp1b1 | CYP450 family 1, subfamily B, polypeptide 1 | NM_009994.1 | 4 | 33↑ | TNX, DN-RTE (3↑) | 65,66 |
| Ccl9 | Chemokine (C-C motif) ligand 9 | NM_011338.2 | 1 | 6↑ | DN-RTE | 65 |
| Clec4d | C-type lectin domain family 4, member d | NM_010819.3 | 1 | 4↑ | DN-RTE | 65 |
| Ctsg | Cathepsin G | NM_007800.1 | 3 | 6↑ | DN-RTE | 65 |
| Ctsl | Cathepsin L | NM_009984.3 | 2 | 4↑ | DN-RTE | 65 |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | NM_009140.2 | 3 | 12↑ | DN-RTE | 65 |
| Fn1 | Fibronectin 1 | NM_010233.1 | 3 | 5↑ | DN-RTE | 65 |
| Hp | Haptoglobin | NM_017370.1 | 2 | 3↑ | DN-RTE | 65 |
| Lpl | Lipoprotein lipase | NM_008509.2 | 2 | 5↑ | DN-RTE | 65 |
| Lyz | Lysozyme | NM_013590.3 | 1 | 3↑ | DN-RTE | 65 |
| Mt1 | Metallothionein 1 | NM_013602.2 | 2 | 3↑ | DN-RTE | 65 |
| S100a8 | S100 calcium binding protein A8 | NM_013650.2 | 1 | 4↑ | DN-RTE | 65 |
| S100a9 | S100 calcium binding protein A9 | NM_009114.2 | 4 | 5↑ | DN-RTE | 65 |
| Scin | Scinderin | NM_009132.1 | 2 | 11↑ | DN-RTE | 65 |
Signalling through the AHR can be down-regulated by at least two means. Upon entering the nucleus, the ligand-activated AHR is exported and degraded by the ubiquitin/proteosome pathway.21 Like many other PAS signalling pathways, AHR signalling includes a negative feedback arm. Signalling by the AHR is attenuated by another PAS protein known as the AHR repressor (AHRR). The AHRR is a DRE-regulated gene and its expression increases rapidly upon AHR activation22 (Fig. 2). The AHRR is structurally similar to the AHR, but contains a potent transcriptional repressor domain and does not require an agonist to dimerize with ARNT. Down-regulation of AHR signalling by these two independent means implies that there is evolutionary selection against overactivation of the AHR gene battery.
AHR-mediated biology
The adaptive pathway
The AHR was originally characterized as a regulator of xenobiotic metabolism, specifically that of polycyclic aromatic hydrocarbons (PAH). Early experiments revealed that exposure to pollutants such as benzo[a]pyrene led to marked increases in cytochromes P450 that act to hydroxylate this foreign chemical, increasing its water solubility and decreasing its biological residency.23 This pathway fits the definition of an ‘adaptive metabolic response’, in that cytosolic AHR binds xenobiotic ligands and activates transcription of enzymes that mediate their biotransformation and excretion.24
The toxic pathway
In response to halogenated dibenzo-p-dioxins (‘dioxins’) and related biphenyls and dibenzofurans, AHR activation not only induces the adaptive xenobiotic metabolic pathway, but also mediates a variety of toxic responses. Dioxin toxicity commonly includes hepatocellular damage, thymic involution, immune suppression, chloracne, epithelial hyperplasia, teratogenesis, and tumour promotion.25–28 Evidence to support the role of the AHR in toxicity is twofold. First, the binding affinity of dioxin congeners for the AHR corresponds to their toxic potency in vivo.29 For example, the ligand TCDD displays the greatest affinity for the AHR and is the most toxic, while the weaker 2,8-dichloro congener has a 300-fold lower affinity and is essentially non-toxic.26,29 Secondly, mice harbouring the Ahrb allele, which codes for a receptor with a high binding affinity for agonists, display an increased incidence of dioxin toxicity and induction of AHR battery genes compared with mice harbouring the Ahrd allele, which encodes a receptor with a 10-fold lower affinity.30–32 In addition to binding affinities with dissociation constant (KD) values in the picomolar range, TCDD is not appreciably metabolized, thus causing prolonged AHR activation.32 Because of its remarkable potency and biological stability, TCDD has proved to be invaluable in elucidating the mechanism of AHR signalling and enzyme induction.
The developmental pathway
To elucidate the physiological and developmental importance of the AHR, several laboratories have characterized Ahr null mice.33–35 These models differ in some respects, yet display key in vivo phenotypic similarities.36 As expected, in response to PAHs and dioxins, Ahr null mice cannot up-regulate the metabolic enzymes characteristic of the adaptive pathway.33–35 Additionally, these animals are resistant to most, if not all, aspects of dioxin toxicity.35 Indicating a role in normal physiology, a number of surprising pathologies also have been characterized in these mouse models. For example, Ahr null mice have markedly smaller livers than wild-type littermates and have abnormal vasculature in the liver, kidney and eye.33,34,37 Abnormal hepatic circulation, characterized by anastomotic sinusoidal vessels, appears to cause decreased perfusion and necrosis of the liver periphery.37,38 This can be demonstrated in Ahr null embryos as early as embryonic day 15.37,38 In addition, extramedullary haematopoiesis, fatty metamorphosis of hepatocytes, and portal tract fibrosis have been observed in Ahr null livers.33,34 Cardiac hypertrophy, hypertension, and elevated levels of vasoconstrictors are also seen in Ahr null animals.39
Perhaps one of the most consistent phenotypic findings in Ahr null animals is a patent ductus venosus (DV).37,40 The DV, like the ductus arteriosus and foramen ovale, is part of the fetal circulatory system.41 The DV connects the umbilical/portal vein to the inferior vena cava, allowing oxygen- and nutrient-rich blood filtered by the placenta to bypass the embryonic liver and nourish the developing heart and brain. Shortly after birth, the DV closes and forces blood from the portal vein through the liver sinusoids for hepatic filtration prior to reaching the lungs and heart. In 100% of Ahr null animals, the DV remains patent into adulthood.40 Because of its robust phenotype–genotype correlation, in our laboratory DV closure is used as a marker of developmental AHR signalling. Hypomorphic Ahr, Arnt, and Ara9 mice, which express only 1/10th of the normal level of protein, also have a patent DV, highlighting the importance of these AHR signalling pathway members in developmental signalling.42–44 Because decreased perfusion and necrosis are seen prior to normal DV closure, the patent DV phenotype in Ahr null animals may be secondary to the abnormal microvascular perfusion. However, the exact mechanism by which the loss of AHR signalling leads to patent DV is still unknown.
Endogenous ligand
The developmental cue for AHR signalling is still unknown. In support of the existence of an ‘endogenous ligand’, we offer the fact that activation of the AHR by TCDD rescues the patent DV phenotype of AHR hypomorphs.43 This observation demonstrates that the developmental response to this toxic ligand mimics developmental AHR signalling. A number of potential endogenous ligands have been suggested. In this regard, indigoids and tryptophan derivatives, which are structurally similar to known xenobiotic ligands, are able to activate AHR signalling.45,46 Recently, low density lipoprotein (LDL) has also been identified as an activator of AHR signalling.47 Because LDL has a very different structure from other known AHR ligands, it is possible that LDL carries into the cell a small molecule with more characteristic AHR agonist features.
However, the developmental cue for AHR activation may not be a ligand at all. Alternative activation mechanisms of the AHR, such as intracellular cyclic AMP (cAMP) and fluid shear stress, have been proposed.47,48 In addition, the AHR may become activated by phosphorylation in response to another cellular cue.49 There is evidence that is inconsistent with the existence of an endogenous ligand. In this regard, mice carrying both the Ahrb and Ahrd alleles, encoding receptors with a 10-fold difference in ligand-binding affinity, display normal DV closure (J. Walisser, unpublished observation). Mouse models have demonstrated that the AHR is required in different cells for toxicity from those required for the developmental role.50 Therefore, the mechanism of TCDD toxicity, clearly linked to ligand binding, is unlikely to be secondary to disruption of the normal developmental role of the AHR. A lack of an endogenous ligand for the AHR would suggest that the adaptive and toxic pathways are independent from the developmental pathway. In fact, there is phylogenetic evidence that the ligand-binding functions of the AHR evolved independently from its developmental role.51
Evolutionary biology of AHR
While the AHR was discovered because of its role in toxicology, the primary function of mammalian AHR is probably related to normal development. Phylogenetic evidence suggests that the vertebrate AHR arose in biological systems over 450 Ma. Therefore, it is unlikely that the products of modern industrialization, PAHs and dioxins, have provided the selective pressure for the conservation of the AHR throughout evolution.51,52 Although nearly all vertebrate AHR orthologues identified to date have been shown to bind TCDD, the response to xenobiotic ligands is quite variable.53,54 The variability in xenobiotic response may have arisen as a means to limit AHR-mediated toxicity while maintaining its key developmental role. Put another way, ligand binding may be a secondary, acquired function of this receptor that arose during vertebrate evolution.
Additional evidence that ligand binding is independent of the developmental role of the AHR comes from data in invertebrate organisms. Invertebrate and vertebrate AHRs share key properties in signal transduction, including heterodimerization with ARNT orthologues and transcriptional activation through DREs.51,53,55,56 However, invertebrate orthologues of the AHR do not bind classic AHR ligands.52,54,55 The DNA sequence of the AHR may have been modified during the evolution of vertebrates, to accommodate an increasing need for AHR signalling in vascular development. These same modifications also may have led to the development of ligand binding.
The vertebrate AHR may function in an analogous manner to the invertebrate AHR but in the vasculature. The Caenorhabditis elegans AHR orthologue directs neuronal cell fate and oxygen-sensitive aggregation.57,58 The Drosophila melanogaster AHR orthologue is expressed in sensory cells and mutations can lead to increased dendritic branching and overgrowth, antennae transforming into legs, and loss of colour vision.58 Although there is little evidence that these same roles of invertebrate AHR have been maintained in mammals, the increased density of anastomotic hepatic sinusoids in the Ahr null mouse is reminiscent of the increased dendritic branching and overgrowth in Drosophila.
AHR in the immune system
AHR-dependent immune function
There is considerable evidence to suggest that AHR signalling plays a role in the function of the immune system. Numerous haematopoietic defects have been described in Ahr null mouse models, including altered lymphocyte numbers in the spleen, perinatal extramedullary haematopoiesis in the liver, and enlarged spleens.33,34 While splenomegaly may be secondary to portal hypertension it may also be a compensation for a haematopoietic defect. In addition to these histological differences, functional studies support the idea that the AHR plays a role in immunity. In this regard, Ahr null animals are more susceptible to listeria infection.59
Considerable evidence from studies using AHR agonists further supports a role for the AHR in the immune system. Exposure to TCDD leads to profound suppression of both humoral and cellular immune responses and results in increased susceptibility to infection.60,61 Although TCDD suppresses CD40L-activated B-cell proliferation, T cells are the primary targets of TCDD and mediate inhibition of the antibody response of B cells.62,63 Thymic involution induced by TCDD is associated with thymocyte loss, thymocyte proliferation arrest and premature emigration of T-cell progenitors.64–66 In addition, TCDD can prevent prothymocytes in the bone marrow from seeding the thymus.67 Three independent laboratories have identified an early triple-negative thymocyte population as the targets of TCDD-induced thymocyte emigration.64–66 Exposure to AHR agonists also affects functional immunity. For example, TCDD causes increased inflammation and inhibits the CD8+ T-cell response to influenza infection.68,69 Other model systems shown to be affected by AHR agonists include experimental autoimmune encephalitis (EAE), graft-versus-host disease (GVHD), and mouse models of allergy and transplant tolerance.70–75
The AHR and Tregs
Recently a role for AHR signalling in regulatory T cells (Tregs) has been reported by at least four independent laboratories. By suppressing effector cell proliferation and cytokine secretion, Tregs have been shown to reduce autoimmune and allergic disease, limit the immune response in infectious disease, and inhibit antitumour immune responses.76 There is evidence that a subset of Tregs develop in the thymus, known as natural or innate Tregs.77 Tregs also develop in the periphery during an immune response and are referred to as adaptive Tregs. These two populations of Tregs probably differ in their antigen specificity, development, and mechanism of immune regulation.78,79 In one report, exposure to TCDD increased the proliferation of Tregs and suppressed EAE.70 In another report, TCDD exposure generated Tregs and prevented GVHD.72 In a third model, activation of the AHR also induced Tregs and improved graft survival.75 In keeping with a role for the AHR in Treg function, it has been observed that naïve T cells isolated from Ahr null animals are inefficient at generating Tregs in vitro.80
The mechanism by which AHR signalling might promote Treg differentiation remains largely uncharacterized. There are many ways to define Tregs. However, there is not yet a clear way to differentiate natural and adaptive Tregs.78 Some laboratories define Tregs by their in vitro suppressive activity or expression of the cell surface markers CD25 and CD62L.72,81 Other laboratories use the expression of forkhead box P3 (FoxP3), a transcription factor thought to play a central role in Treg activity.70,80 Exposure to TCDD causes a reduction in CD62L expression in T cells.82 There is evidence that the AHR directly regulates the expression of FoxP3, and AHR activation leads to an increase in Tregs in at least two model systems.70,75,80 However, a decrease in FoxP3+ cells upon activation of the AHR was seen in a GVHD model.72 This inconsistency in the effect of AHR signalling on FoxP3+ cells may be explained by the dose of TCDD or the immune response model used in the experiments. The variable effect on FoxP3 expression also may suggest that AHR signalling plays different roles in natural and adaptive Tregs and that the type of Tregs involved in EAE, allograft-tolerance and GVHD models differs.
The AHR may affect Treg differentiation through at least two other mechanisms. First, AHR signalling may influence Treg development by augmenting transforming growth factor (TGF)-β signalling. A number of independent laboratories using a variety of ligands and experimental systems have identified an interplay between the AHR and TGF-β signalling pathways.83–86 Cross-talk between AHR and TGF-β signalling also has been observed during Treg differentiation. For example, a 13-fold increase in TGF-β3 RNA was found in Tregs exposed to TCDD.72 Furthermore, in tissue culture, TGF-β mimics the effects of TCDD on Tregs, and inhibition of TGF-β signalling also inhibits TCDD-induced Treg activity.70 It has been suggested that the Treg populations have different requirements for TGF-β.78 In fact, there are fewer peripheral Tregs, but normal numbers of thymus-derived Tregs, in Tgfbr null mice.87 These data also provide evidence that the AHR may play different roles in adaptive and natural Tregs.
Another potential mechanism by which the AHR may affect Treg activity involves dendritic cells (DCs). DC antigen presentation plays a central role in converting naïve T cells into Tregs in the periphery (adaptive Tregs).88,89 Cytokines are crucial to T-cell activation, and without the appropriate milieu, DCs can induce clonal deletion, anergy or tolerogenic regulatory T cells.90,91 A model exists whereby mature DCs activate Tregs, and these Tregs go on to limit their own expansion by blocking splenic DCs from maturing.89 Exposure to TCDD reduces the number of splenic DCs.92 The AHR agonist VAF347 promoted long-term graft acceptance in an islet cell transplant model and reduced the response in an allergy model.74,75 In this study, alterations in DC expression of interleukin (IL)-6, IL-10 and TGF-β were proposed as a potential mechanism of the AHR-mediated effect. In fact, the authors demonstrated that graft rejection was prevented by transfer of AHR agonist-treated DCs. Taken together, these findings suggest that AHR signalling may affect Treg differentiation by modulating expression of Treg markers from within Tregs or through DCs.
AHR-mediated inflammation
The AHR has been reported recently to play a role in the development of T helper 17 (Th17) cells, a new subset of CD4+ T cells thought to play a major role in autoimmunity and clearance of infectious agents. It has been observed that injecting healthy mice with Th17 cells from animals with EAE causes autoimmunity in the recipients.93 In addition, it has been shown that adequate Th17 cell function inhibits systemic infection with gastrointestinal pathogens.93,94 Th17 cells are characterized by their secretion of the proinflammatory cytokines IL-17 and IL-22. The ligand-activated AHR regulates expression of these cytokines in tissue culture.71 A role for the AHR in the regulation of Th17 cells is supported further by the observation that the absolute number of Th17 cells is reduced in Ahr null mice upon induction of EAE.71,80
Because Th17 cells promote the immune response and Tregs are known to decrease immune reactivity, a model has emerged suggesting that the Treg/Th17 balance distinguishes an effective immune response and self-antigen tolerance from chronic infection or autoimmunity. Preliminary evidence from multiple laboratories has suggested that the AHR modifies the Treg/Th17 cell balance through modifying the cytokine milleu. The mechanism at work may be related to the fact that TGF-β induces Treg differentiation, while the presence of IL-6 leads to TGF-β-dependent Th17 cell production.76,93 As described above, the AHR has been shown to modulate cytokine signalling. Further evidence for this biology is provided by unpublished observations from our own laboratory, where we observed that Helicobacter infection results in rectal prolapse in Ahr null animals, in marked contrast to wild-type littermates (E. Stevens, unpublished observation). It is possible that an alteration in the Treg/Th17 balance in Ahr null animals may play a role in the severity of this gastrointestinal infection.
As a transcription factor, AHR probably modulates T-cell development at the transcriptional level. In addition to subtype-specific transcriptional changes described above, the up-regulation of CD11a is blocked in activated T cells treated with TCDD, which may impede T cells from reaching the source of antigen.82 As chemokine receptor transcripts are up-regulated, T cells disappear from the spleen, suggesting homing to other tissues.72 It has also been demonstrated that TCDD up-regulates Kruppel like factor 2 (Klf2), which is implicated in the homing of T cells and the prevention of inflammatory chemokine receptor expression.66,95 In summary, the published data to date are in agreement that AHR-mediated transcriptional changes can affect T-cell activation, but the mechanism is still largely unknown.
Exposure to TCDD can lead to inflammation and also to increased secretion of inflammatory cytokines involved in innate immunity.28 In mice, TCDD exposure is associated with decreased survival, neutrophilia, and elevated interferon (IFN)-γ levels in the lungs following influenza virus infection.68 Given that AHR in haematopoietic cells is not required for this phenotype, it may be that activation within the lung parenchyma changes the immune response to infection.68,96 One interpretation of this data is that neutrophils are a secondary response to AHR-mediated transcriptional changes within the lung parenchymal target cell. This model may be similar to what is occurring in TCDD-induced hepatotoxicity. In this system, conditional AHR mouse models have been used to demonstrate that TCDD causes primary transcriptional effects within hepatocytes and secondary effects are mediated by inflammatory cells that exacerbate the hepatotoxicity.97 These experiments emphasize the importance of determining the primary effects of AHR signalling in order to elucidate the mechanism by which AHR signalling affects the immune response.
Considerations for future experiments
Primary and secondary effects in AHR biology
We propose a model in which all the upstream signalling steps in the three AHR biological pathways are similar. To support this idea, we offer the fact that both TCDD-induced toxicity and vascular development are dependent on most, if not all, of the same signalling molecules required for the adaptive response.42,44 Given the importance of enhancer recognition of the AHR:ARNT heterodimer, it would follow that transcriptional changes are the primary mechanism leading to both developmental and toxic end-points. In support of this idea, AHR mutants deficient in DNA-binding or nuclear localization activity are also resistant to TCDD-induced toxicity and display patent DV.98,99 In our view, these experiments suggest that elucidating the specific transcriptional targets of AHR underlying each distinct biological end-point is critical to defining the mechanism.
One key point is that, while the signalling mechanism of gene transcription is central to all AHR biology, which genes are targeted depends largely on the cell type being studied. Data demonstrating that the AHR is required in different cell types for hepatotoxicity and vascular development are in agreement with the idea that the AHR mediates various roles in a cell type-dependent fashion.50 Therefore, identifying the target cell(s) is an important first step in the identification of key gene targets mediating such variable end-points as hepatoxicity, vascular remodelling, and immune suppression in AHR biology.
Another key point to consider when studying the mechanism by which the AHR mediates such diverse biology is the distinction between primary and downstream gene targets. While DRE-driven, AHR-mediated transcription has been well studied, DRE-independent gene transcription has also been reported to occur. These DRE-independent targets may represent promoters bound by the AHR at enhancer sequences distinct from DREs or they may be secondary targets.100 Using dose response and timing of transcriptional changes, the primary genetic targets of AHR activation in the liver have been separated from the downstream transcriptional changes during TCDD-induced hepatoxicity.101,102 Although many AHR target genes have been identified, their position in the signalling sequence and the mechanism by which they mediate AHR biology still must be elucidated (Table 1). In conclusion, identifying the primary transcriptional changes mediated by the AHR in target cells holds promise for elucidation of the role of AHR in the immune system.
AHR pharmacology
The well-characterized pharmacology of the AHR may prove to be a powerful tool with which to unravel the role of this protein in immunology. The most studied AHR agonist, TCDD, binds AHR with high affinity.103,104 Because its four chlorine residues prevent access to the active sites of metabolic enzymes, TCDD is poorly metabolized.103 As a relatively pure, high-affinity agonist, TCDD can be used at low doses and thus would be predicted to have fewer off-target effects than non-halogenated agonists. For example, indirubin is a potent AHR agonist, but it also binds to and inhibits cyclin-dependent kinases and c-Src kinase.105 Benzo-a-pyrene activates the AHR and is metabolized to epoxides and quinones via the adaptive pathway.106 Epoxide intermediates are known to be highly cytotoxic through alkylation of DNA and other cellular macromolecules.106 Quinones can generate reactive oxygen species (ROS), which in turn can influence gene expression through a variety of mechanisms, including activation of the transcription factor nuclear erythroid 2-related factor (Nrf2).106,107 The bottom line is that studies using highly potent and specific agonists, such as TCDD, provide the most direct route to elucidate the signalling steps by which the AHR influences the immune system.
Another important pharmacological consideration in the study of AHR signalling is the length of activation. Unlike TCDD, many AHR agonists, including benzo-a-pyrene (BaP), indirubin and 6-formylindolo(3,2-b) carbazole (FICZ), are rapidly cleared, leading to only short-term activation.103 These agonists cause substantial up-regulation of AHR battery genes, but only within hours of treatment.103,108 In comparison, TCDD causes long-term stimulation of AHR that can be measured days after administration.109–111 Differences in the length of AHR stimulation may lead to differences in AHR-mediated biology. In fact, data from experiments using FICZ and TCDD led to different conclusions about AHR signalling in T cells during EAE.70 It is clear that the choice of agonist is an important consideration for the design and interpretation of future studies (Table 2).
Table 2.
Aryl hydrocarbon receptor (AHR) ligands
| Ligand | EC50 (mol/kg) | Metabolized? | References |
|---|---|---|---|
| HAH (TCDD) | 10−12 | No | 116,117 |
| PAH (BaP) | 10−5–10−6 | Yes | 117,118 |
| PCB (pentaCB) | 10−7 | Yes | 104 |
| FICZ | 10−10–10−12 | Yes | 109,119 |
| Indirubin | 10−8 | Yes | 45,120 |
| Lipoxin-4a | 10−9 | Yes | 121 |
| Bilirubin | 10−6 | Yes | 122 |
| ITE | 10−9 | ? | 123,124 |
| ICZ | 10−8–10−10 | Yes | 116,125 |
This table contains some of the best studied AHR ligands. The EC50 is the dose of the ligand that leads to 50% of the maximal cytochrome P450 gene induction. These are estimates and are dependent on many factors, including cell type and AHR allele. Some AHR ligands are metabolized enzymatically and are short-lived. The EC50s of these ligands are sensitive to the timing of induction.
FICZ, 6-formylindolo(3,2-b)carbazole; ICZ, indolo(3,2-b)carbazole; ITE, 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester; PAH, polycyclic aromatic hydrocarbon; PCB, polychlorinated biphenyl; TCDD, 2,3,7,8 tetrachlorodibenzo-p-dioxin.
Use of Ahr mutant animals
The use of Ahr null animals complicates AHR pharmacology. There are two reasons why the use of Ahr null animals can lead to misinterpretation of results. First, these mice have a patent DV and other abnormalities described above. It can, therefore, be difficult to isolate phenotypic effects directly caused by the loss of the AHR and those downstream of abnormal vasculature. Secondly, Ahr null animals should not be used to study ligands that may be modified by metabolic enzymes. Bioactivation may be required for the phenotypic effect. The use of Ahr null animals in these situations does not allow the conclusion that the phenotype is the direct result of an AHR transcriptional response, as loss of the AHR impairs metabolic enzyme induction. To test the requirement for the AHR in the pharmacology of any compound, it is imperative to create mice with the Ahr deleted specifically in a target cell subset, to avoid patent DV and its resulting pathologies. Such a mouse model can be created with bone marrow chimeras or with the conditional Ahrfx allele and cre recombinase mediated LoxP site recombination (Creû–Lox) technology.
Conclusion
Much has been elucidated about AHR biology in the last 30 years. It is clear that this receptor is not simply a transcription factor developed to respond to toxicants, but probably plays a central role in vascular biology. Activation of this receptor has long been known to cause immunosuppression and thymic involution. Recent data have implicated this receptor in T-cell differentiation and DC function. There is little dispute that T-cell lineage specificity is largely determined by transcription factors and that gene transcription plays large roles in immune responses in other haematopoietic lineages. As a transcription factor, the AHR probably modulates immune reactions and also causes immunotoxicity through transcriptional changes. It seems likely that a role for this receptor in the function of the immune system will be defined.
Acknowledgments
Special thanks to B. E. McIntosh for critical reading of this manuscript. EAS is supported by an NRSA from the National Institutes of Health (NIH; Grants F30-ES015416). This work was supported by NIH grants ES005703 and P30-CA014520.
Glossary
Abbreviations:
- AHR
aryl hydrocarbon receptor
- AHRR
aryl hydrocarbon receptor repressor
- ARA9
aryl hydrocarbon receptor associated 9
- ARNT
aryl hydrocarbon receptor nuclear translocator
- BaP
benzo-a-pyrene
- bHLH
basic helix loop helix
- Cre-Lox
cre recombinase mediated LoxP site recombination
- CYP
cytochrome P450
- DC
dendritic cell
- DRE
dioxin response element
- EAE
experimental autoimmune encephalitis
- FICZ
6-formylindolo(3,2-b)carbazole
- FoxP3
forkhead box P3
- GVHD
graft versus host disease
- HAH
halogenated aryl hydrocarbon
- Hsp90
heat shock protein 90
- ICZ
indolo(3,2-b)carbazole
- ITE
2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester
- KD
dissociation constant
- Klf2
Kruppel like factor 2
- LDL
low density lipoprotein
- NES
nuclear export signal
- NLS
nuclear localization signal
- Nrf2
nuclear erythroid 2-related factor
- p23
protein 23
- PAH
polycyclic aromatic hydrocarbon
- PAS
Per-ARNT-Sim
- PCB
polychlorinated biphenyl
- pentaCB
penta chlorinated byphenyls
- RIP140
receptor interacting protein 140
- SHP
nuclear receptor subfamily 0 group B member 2
- Sp1
transacting transcription factor 1
- SRC
nuclear receptor coactivator
- TAD
transcriptionally active domain
- TCDD
2,3,7,8 tetrachlorodibenzo-p-dioxin
- Th17
T-helper 17
- Treg
regulatory T cell
- UGT1A6
UDP glucuronosyltransferase 1 family polypeptide A6
Disclosures
CAB has served as a consultant to Dow Chemical Company on issues related to dioxin toxicology.
References
- 1.Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci USA. 1997;94:13743–8. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–61. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 3.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci USA. 1992;89:8185–9. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolwick KM, Schmidt JV, Carver LA, Swanson HI, Bradfield CA. Cloning and expression of a human Ah receptor cDNA. Mol Pharmacol. 1993;44:911–17. [PubMed] [Google Scholar]
- 5.Dolwick KM, Swanson HI, Bradfield CA. In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc Natl Acad Sci USA. 1993;90:8566–70. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem. 1995;270:29270–8. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 7.Coumailleau P, Poellinger L, Gustafsson JA, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 8.Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 9.Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267:13728–34. [PubMed] [Google Scholar]
- 10.Kazlauskas A, Poellinger L, Pongratz I. The immunophilin-like protein XAP2 regulates ubiquitination and subcellular localization of the dioxin receptor. J Biol Chem. 2000;275:41317–24. doi: 10.1074/jbc.M007765200. [DOI] [PubMed] [Google Scholar]
- 11.Kazlauskas A, Sundstrom S, Poellinger L, Pongratz I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Molecular & Cellular Biology. 2001;21(7):2594–607. doi: 10.1128/MCB.21.7.2594-2607.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem. 1998;273:33580–7. doi: 10.1074/jbc.273.50.33580. [DOI] [PubMed] [Google Scholar]
- 13.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–5. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 14.Yao G, Craven M, Drinkwater N, Bradfield CA. Interaction networks in yeast define and enumerate the signaling steps of the vertebrate aryl hydrocarbon receptor. PLoS Biol. 2004;2:E65. doi: 10.1371/journal.pbio.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beischlag TV, Wang S, Rose DW, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22:4319–33. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J Biochem. 1997;122:703–10. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- 17.Lemon BD, Freedman LP. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen TA, Hoivik D, Lee JE, Safe S. Interactions of nuclear receptor coactivator/corepressor proteins with the aryl hydrocarbon receptor complex. Arch Biochem Biophys. 1999;367:250–7. doi: 10.1006/abbi.1999.1282. [DOI] [PubMed] [Google Scholar]
- 19.Klinge CM, Jernigan SC, Risinger KE, Lee JE, Tyulmenkov VV, Falkner KC, Prough RA. Short heterodimer partner (SHP) orphan nuclear receptor inhibits the transcriptional activity of aryl hydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT) Arch Biochem Biophys. 2001;390:64–70. doi: 10.1006/abbi.2001.2366. [DOI] [PubMed] [Google Scholar]
- 20.Nebert DW, Puga A, Vasiliou V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann N Y Acad Sci. 1993;685:624–40. doi: 10.1111/j.1749-6632.1993.tb35928.x. [DOI] [PubMed] [Google Scholar]
- 21.Davarinos NA, Pollenz RS. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J Biol Chem. 1999;274:28708–15. doi: 10.1074/jbc.274.40.28708. [DOI] [PubMed] [Google Scholar]
- 22.Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 1999;13:20–5. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes memorial lecture. Cancer Res. 1982;42:4875–917. [PubMed] [Google Scholar]
- 24.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 25.Tomita S, Jiang HB, Ueno T, et al. T cell-specific disruption of arylhydrocarbon receptor nuclear translocator (Arnt) gene causes resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced thymic involution. J Immunol. 2003;171:4113–20. doi: 10.4049/jimmunol.171.8.4113. [DOI] [PubMed] [Google Scholar]
- 26.Schecter A, Birnbaum L, Ryan JJ, Constable JD. Dioxins: an overview. Environ Res. 2006;101:419–28. doi: 10.1016/j.envres.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Pitot HC, Goldsworthy T, Campbell HA, Poland A. Quantitative evaluation of the promotion by 2,3,7,8-tetrachlorodibenzo-p-dioxin of hepatocarcinogenesis from diethylnitrosamine. Cancer Res. 1980;40:3616–20. [PubMed] [Google Scholar]
- 28.Holsapple MP, Morris DL, Wood SC, Snyder NK. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced changes in immunocompetence: possible mechanisms. Annu Rev Pharmacol Toxicol. 1991;31:73–100. doi: 10.1146/annurev.pa.31.040191.000445. [DOI] [PubMed] [Google Scholar]
- 29.Safe SH. Comparative toxicology and mechanism of action of polychlorinated dibenzo-p-dioxins and dibenzofurans. Annu Rev Pharmacol Toxicol. 1986;26:371–99. doi: 10.1146/annurev.pa.26.040186.002103. [DOI] [PubMed] [Google Scholar]
- 30.Thomas RS, Rank DR, Penn SG, et al. Application of genomics to toxicology research. Environ Health Perspect. 2002;110(Suppl 6):919–23. doi: 10.1289/ehp.02110s6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poland A, Glover E. 2,3,7,8,-Tetrachlorodibenzo-p-dioxin: segregation of toxocity with the Ah locus. Mol Pharmacol. 1980;17:86–94. [PubMed] [Google Scholar]
- 32.Poland A, Palen D, Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol Pharmacol. 1994;46:915–21. [PubMed] [Google Scholar]
- 33.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA. 1996;93:6731–6. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Salguero P, Pineau T, Hilbert DM, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–6. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 35.Mimura J, Yamashita K, Nakamura K, et al. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes to Cells. 1997;2:645–54. doi: 10.1046/j.1365-2443.1997.1490345.x. [DOI] [PubMed] [Google Scholar]
- 36.Lahvis GP, Bradfield CA. Ahr null alleles: distinctive or different? Biochem Pharmacol. 1998;56:781–7. doi: 10.1016/s0006-2952(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 37.Lahvis G, Lindell S, Thomas R, et al. Portosystemic shunts and persistent fetal vascular structures in Ah-receptor deficient mice. Proc Natl Acad Sci USA. 2000;97:10442–7. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harstad EB, Guite CA, Thomae TL, Bradfield CA. Liver deformation in Ahr-null mice: evidence for aberrant hepatic perfusion in early development. Mol Pharmacol. 2006;69:1534–41. doi: 10.1124/mol.105.020107. [DOI] [PubMed] [Google Scholar]
- 39.Lund AK, Goens MB, Kanagy NL, Walker MK. Cardiac hypertrophy in aryl hydrocarbon receptor null mice is correlated with elevated angiotensin II, endothelin-1, and mean arterial blood pressure. Toxicol Appl Pharmacol. 2003;193:177–87. doi: 10.1016/j.taap.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol. 2005;67:714–20. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- 41.Kiserud T. Physiology of the fetal circulation. Semin Fetal Neonatal Med. 2005;10:493–503. doi: 10.1016/j.siny.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Walisser JA, Bunger MK, Glover E, Harstad EB, Bradfield CA. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J Biol Chem. 2004;279:16326–31. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- 43.Walisser JA, Bunger MK, Glover E, Bradfield CA. Gestational exposure of Ahr and Arnt hypomorphs to dioxin rescues vascular development. Proc Natl Acad Sci USA. 2004;101:16677–82. doi: 10.1073/pnas.0404379101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin BC, Nguyen LP, Walisser JA, Bradfield CA. A hypomorphic allele of aryl hydrocarbon receptor-associated protein-9 produces a phenocopy of the AHR-null mouse. Mol Pharmacol. 2008;74:1367–71. doi: 10.1124/mol.108.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guengerich FP, Martin MV, McCormick WA, Nguyen LP, Glover E, Bradfield CA. Aryl hydrocarbon receptor response to indigoids in vitro and in vivo. Arch Biochem Biophys. 2004;423:309–16. doi: 10.1016/j.abb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Helferich WG, Denison MS. Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol Pharmacol. 1991;40:674–8. [PubMed] [Google Scholar]
- 47.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor is activated by modified low-density lipoprotein. Proc Natl Acad Sci USA. 2007;104:1412–17. doi: 10.1073/pnas.0607296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oesch-Bartlomowicz B, Huelster A, Wiss O, et al. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proc Natl Acad Sci USA. 2005;102:9218–23. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puga A, Marlowe J, Barnes S, et al. Role of the aryl hydrocarbon receptor in cell cycle regulation. Toxicology. 2002;182:171–7. doi: 10.1016/s0300-483x(02)00276-7. [DOI] [PubMed] [Google Scholar]
- 50.Walisser JA, Glover E, Pande K, Liss A, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci USA. 2005;102:17858–63. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn M. Aryl hydrocarbon receptors: diversity and evolution(1) Chem Biol Interact. 2002;141:131. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 52.Hahn ME, Poland A, Glover E, Stegeman JJ. Photoaffinity labeling of the Ah receptor: phylogenetic survey of diverse vertebrate and invertebrate species. Arch Biochem Biophys. 1994;310:218–28. doi: 10.1006/abbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 53.Thomas RS, Penn SG, Holden K, Bradfield CA, Rank DR. Sequence variation and phylogenetic history of the mouse Ahr gene. Pharmacogenetics. 2002;12:151–63. doi: 10.1097/00008571-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 54.Hahn ME, Karchner SI, Evans BR, Franks DG, Merson RR, Lapseritis JM. Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: insights from comparative genomics. J Exp Zoolog A Comp Exp Biol. 2006;305:693–706. doi: 10.1002/jez.a.323. [DOI] [PubMed] [Google Scholar]
- 55.Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci USA. 1998;95:2844–9. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emmons RB, Duncan D, Estes PA, et al. The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–45. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- 57.Qin H, Zhai Z, Powell-Coffman JA. The Caenorhabditis elegans AHR-1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev Biol. 2006;298:606–15. doi: 10.1016/j.ydbio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 58.Crews ST, Brenman JE. Spineless provides a little backbone for dendritic morphogenesis. Genes Dev. 2006;20:2773–8. doi: 10.1101/gad.1487706. [DOI] [PubMed] [Google Scholar]
- 59.Shi LZ, Faith NG, Nakayama Y, Suresh M, Steinberg H, Czuprynski CJ. The aryl hydrocarbon receptor is required for optimal resistance to Listeria monocytogenes infection in mice. J Immunol. 2007;179:6952–62. doi: 10.4049/jimmunol.179.10.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerkvliet NI, Baecher-Steppan L, Shepherd DM, Oughton JA, Vorderstrasse BA, DeKrey GK. Inhibition of TC-1 cytokine production, effector cytotoxic T lymphocyte development and alloantibody production by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Immunol. 1996;157:2310–19. [PubMed] [Google Scholar]
- 61.Kerkvliet NI. Immunological effects of chlorinated dibenzo-p-dioxins. Environ Health Perspect. 1995;103(Suppl 9):47–53. doi: 10.1289/ehp.95103s947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allan LL, Sherr DH. Constitutive activation and environmental chemical induction of the aryl hydrocarbon receptor/transcription factor in activated human B lymphocytes. Mol Pharmacol. 2005;67:1740–50. doi: 10.1124/mol.104.009100. [DOI] [PubMed] [Google Scholar]
- 63.Ito T, Inouye K, Fujimaki H, Tohyama C, Nohara K. Mechanism of TCDD-induced suppression of antibody production: effect on T cell-derived cytokine production in the primary immune reaction of mice. Toxicol Sci. 2002;70:46–54. doi: 10.1093/toxsci/70.1.46. [DOI] [PubMed] [Google Scholar]
- 64.Laiosa MD, Wyman A, Murante FG, Fiore NC, Staples JE, Gasiewicz TA, Silverstone AE. Cell proliferation arrest within intrathymic lymphocyte progenitor cells causes thymic atrophy mediated by the aryl hydrocarbon receptor. J Immunol. 2003;171:4582–91. doi: 10.4049/jimmunol.171.9.4582. [DOI] [PubMed] [Google Scholar]
- 65.Temchura VV, Frericks M, Nacken W, Esser C. Role of the aryl hydrocarbon receptor in thymocyte emigration in vivo. Eur J Immunol. 2005;35:2738–47. doi: 10.1002/eji.200425641. [DOI] [PubMed] [Google Scholar]
- 66.McMillan BJ, McMillan SN, Glover E, Bradfield CA. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces premature activation of the KLF2 regulon during thymocyte development. J Biol Chem. 2007;282:12590–7. doi: 10.1074/jbc.M611446200. [DOI] [PubMed] [Google Scholar]
- 67.Fine JS, Silverstone AE, Gasiewicz TA. Impairment of prothymocyte activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Immunol. 1990;144:1169–76. [PubMed] [Google Scholar]
- 68.Neff-LaFord H, Teske S, Bushnell TP, Lawrence BP. Aryl hydrocarbon receptor activation during influenza virus infection unveils a novel pathway of IFN-gamma production by phagocytic cells. J Immunol. 2007;179:247–55. doi: 10.4049/jimmunol.179.1.247. [DOI] [PubMed] [Google Scholar]
- 69.Mitchell KA, Lawrence BP. Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) renders influenza virus-specific CD8+ T cells hyporesponsive to antigen. Toxicol Sci. 2003;74:74–84. doi: 10.1093/toxsci/kfg110. [DOI] [PubMed] [Google Scholar]
- 70.Quintana FJ, Basso AS, Iglesias AH, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 71.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–9. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 72.Marshall NB, Vorachek WR, Steppan LB, Mourich DV, Kerkvliet NI. Functional characterization and gene expression analysis of CD4+ CD25+ regulatory T cells generated in mice treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Immunol. 2008;181:2382–91. doi: 10.4049/jimmunol.181.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kerkvliet NI, Shepherd DM, Baecher-Steppan L. T lymphocytes are direct, aryl hydrocarbon receptor (AhR)-dependent targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): AhR expression in both CD4+ and CD8+ T cells is necessary for full suppression of a cytotoxic T lymphocyte response by TCDD. Toxicol Appl Pharmacol. 2002;185:146–52. doi: 10.1006/taap.2002.9537. [DOI] [PubMed] [Google Scholar]
- 74.Lawrence BP, Denison MS, Novak H, Vorderstrasse BA, Harrer N, Neruda W, Reichel C, Woisetschlager M. Activation of the aryl hydrocarbon receptor is essential for mediating the anti-inflammatory effects of a novel low-molecular-weight compound. Blood. 2008;112:1158–65. doi: 10.1182/blood-2007-08-109645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hauben E, Gregori S, Draghici E, Migliavacca B, Olivieri S, Woisetschlager M, Roncarolo MG. Activation of the aryl hydrocarbon receptor promotes allograft-specific tolerance through direct and dendritic cell-mediated effects on regulatory T cells. Blood. 2008;112:1214–22. doi: 10.1182/blood-2007-08-109843. [DOI] [PubMed] [Google Scholar]
- 76.Mottet C, Golshayan D. CD4+ CD25+ Foxp3+ regulatory T cells: from basic research to potential therapeutic use. Swiss Med Wkly. 2007;137:625–34. doi: 10.4414/smw.2007.11916. [DOI] [PubMed] [Google Scholar]
- 77.Afzali B, Lombardi G, Lechler RI, Lord GM. The role of T helper 17 (Th17) and regulatory T cells (Treg) in human organ transplantation and autoimmune disease. Clin Exp Immunol. 2007;148:32–46. doi: 10.1111/j.1365-2249.2007.03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You S, Leforban B, Garcia C, Bach JF, Bluestone JA, Chatenoud L. Adaptive TGF-beta-dependent regulatory T cells control autoimmune diabetes and are a privileged target of anti-CD3 antibody treatment. Proc Natl Acad Sci USA. 2007;104:6335–40. doi: 10.1073/pnas.0701171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nat Rev Immunol. 2003;3:253–7. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 80.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci USA. 2008;105:9721–6. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Funatake CJ, Marshall NB, Steppan LB, Mourich DV, Kerkvliet NI. Cutting edge: activation of the aryl hydrocarbon receptor by 2,3,7,8-tetrachlorodibenzo-p-dioxin generates a population of CD4+ CD25+ cells with characteristics of regulatory T cells. J Immunol. 2005;175:4184–8. doi: 10.4049/jimmunol.175.7.4184. [DOI] [PubMed] [Google Scholar]
- 82.Funatake CJ, Dearstyne EA, Steppan LB, Shepherd DM, Spanjaard ES, Marshak-Rothstein A, Kerkvliet NI. Early consequences of 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure on the activation and survival of antigen-specific T cells. Toxicol Sci. 2004;82:129–42. doi: 10.1093/toxsci/kfh245. [DOI] [PubMed] [Google Scholar]
- 83.Santiago-Josefat B, Mulero-Navarro S, Dallas SL, Fernandez-Salguero PM. Overexpression of latent transforming growth factor-beta binding protein 1 (LTBP-1) in dioxin receptor-null mouse embryo fibroblasts. J Cell Sci. 2004;117(6):849–59. doi: 10.1242/jcs.00932. Pt. [DOI] [PubMed] [Google Scholar]
- 84.Gomez-Duran A, Mulero-Navarro S, Chang X, Fernandez-Salguero PM. LTBP-1 blockade in dioxin receptor-null mouse embryo fibroblasts decreases TGF-beta activity: role of extracellular proteases plasmin and elastase. J Cell Biochem. 2006;97:380–92. doi: 10.1002/jcb.20637. [DOI] [PubMed] [Google Scholar]
- 85.Thomae TL, Stevens EA, Bradfield CA. Transforming growth factor-beta3 restores fusion in palatal shelves exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 2005;280:12742–6. doi: 10.1074/jbc.M410780200. [DOI] [PubMed] [Google Scholar]
- 86.Guo J, Sartor M, Karyala S, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Expression of genes in the TGF-beta signaling pathway is significantly deregulated in smooth muscle cells from aorta of aryl hydrocarbon receptor knockout mice. Toxicol Appl Pharmacol. 2004;194:79–89. doi: 10.1016/j.taap.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Huber S, Schramm C, Lehr HA, et al. Cutting edge: TGF-beta signaling is required for the in vivo expansion and immunosuppressive capacity of regulatory CD4+ CD25+ T cells. J Immunol. 2004;173:6526–31. doi: 10.4049/jimmunol.173.11.6526. [DOI] [PubMed] [Google Scholar]
- 88.Thorstenson KM, Khoruts A. Generation of anergic and potentially immunoregulatory CD25+ CD4 T cells in vivo after induction of peripheral tolerance with intravenous or oral antigen. J Immunol. 2001;167:188–95. doi: 10.4049/jimmunol.167.1.188. [DOI] [PubMed] [Google Scholar]
- 89.Yamazaki S, Inaba K, Tarbell KV, Steinman RM. Dendritic cells expand antigen-specific Foxp3+ CD25+ CD4+ regulatory T cells including suppressors of alloreactivity. Immunol Rev. 2006;212:314–29. doi: 10.1111/j.0105-2896.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- 90.Miller SD, Turley DM, Podojil JR. Antigen-specific tolerance strategies for the prevention and treatment of autoimmune disease. Nat Rev Immunol. 2007;7:665–77. doi: 10.1038/nri2153. [DOI] [PubMed] [Google Scholar]
- 91.Akbari O, Freeman GJ, Meyer EH, et al. Antigen-specific regulatory T cells develop via the ICOS-ICOS-ligand pathway and inhibit allergen-induced airway hyperreactivity. Nat Med. 2002;8:1024–32. doi: 10.1038/nm745. [DOI] [PubMed] [Google Scholar]
- 92.Vorderstrasse BA, Kerkvliet NI. 2,3,7,8-Tetrachlorodibenzo-p-dioxin affects the number and function of murine splenic dendritic cells and their expression of accessory molecules. Toxicol Appl Pharmacol. 2001;171:117–25. doi: 10.1006/taap.2000.9119. [DOI] [PubMed] [Google Scholar]
- 93.Larosa DF, Orange JS. 1. Lymphocytes. J Allergy Clin Immunol. 2008;2(Suppl):S364–9. doi: 10.1016/j.jaci.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 94.O’Keeffe J, Moran AP. Conventional, regulatory, and unconventional T cells in the immunologic response to Helicobacter pylori. Helicobacter. 2008;13:1–19. doi: 10.1111/j.1523-5378.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 95.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 96.Teske S, Bohn AA, Hogaboam JP, Lawrence BP. Aryl hydrocarbon receptor targets pathways extrinsic to bone marrow cells to enhance neutrophil recruitment during influenza virus infection. Toxicol Sci. 2008;102:89–99. doi: 10.1093/toxsci/kfm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pande K, Moran SM, Bradfield CA. Aspects of dioxin toxicity are mediated by interleukin 1-like cytokines. Mol Pharmacol. 2005;67:1393–8. doi: 10.1124/mol.105.010983. [DOI] [PubMed] [Google Scholar]
- 98.Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–74. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 99.Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, Bradfield CA. Abnormal Liver Development and Resistance to 2,3,7,8-Tetrachlorodibenzo-p-dioxin Toxicity in Mice Carrying a Mutation in the DNA Binding Domain of the Aryl Hydrocarbon Receptor. Toxicol Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boutros PC, Moffat ID, Franc MA, Tijet N, Tuomisto J, Pohjanvirta R, Okey AB. Dioxin-responsive AHRE-II gene battery: identification by phylogenetic footprinting. Biochem Biophys Res Commun. 2004;321:707–15. doi: 10.1016/j.bbrc.2004.06.177. [DOI] [PubMed] [Google Scholar]
- 101.Hayes KR, Zastrow GM, Nukaya M, et al. Hepatic transcriptional networks induced by exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem Res Toxicol. 2007;20:1573–81. doi: 10.1021/tx7003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ovando BJ, Vezina CM, McGarrigle BP, Olson JR. Hepatic gene downregulation following acute and subchronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2006;94:428–38. doi: 10.1093/toxsci/kfl111. [DOI] [PubMed] [Google Scholar]
- 103.Bohonowych JE, Denison MS. Persistent binding of ligands to the aryl hydrocarbon receptor. Toxicol Sci. 2007;98:99–109. doi: 10.1093/toxsci/kfm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit Rev Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 105.Sethi G, Ahn KS, Sandur SK, Lin X, Chaturvedi MM, Aggarwal BB. Indirubin enhances tumor necrosis factor-induced apoptosis through modulation of nuclear factor-kappa B signaling pathway. J Biol Chem. 2006;281:23425–35. doi: 10.1074/jbc.M602627200. [DOI] [PubMed] [Google Scholar]
- 106.Burchiel SW, Luster MI. Signaling by environmental polycyclic aromatic hydrocarbons in human lymphocytes. Clin Immunol. 2001;98:2–10. doi: 10.1006/clim.2000.4934. [DOI] [PubMed] [Google Scholar]
- 107.Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–43. doi: 10.5483/bmbrep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- 108.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–16. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oberg M, Bergander L, Hakansson H, Rannug U, Rannug A. Identification of the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole, in cell culture medium, as a factor that controls the background aryl hydrocarbon receptor activity. Toxicol Sci. 2005;85:935–43. doi: 10.1093/toxsci/kfi154. [DOI] [PubMed] [Google Scholar]
- 110.Morales JL, Krzeminski J, Amin S, Perdew GH. Characterization of the antiallergic drugs 3-[2-(2-phenylethyl) benzoimidazole-4-yl]-3-hydroxypropanoic acid and ethyl 3-hydroxy-3-[2-(2-phenylethyl)benzoimidazol-4-yl]propanoate as full aryl hydrocarbon receptor agonists. Chem Res Toxicol. 2008;21:472–82. doi: 10.1021/tx700350v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Negishi T, Kato Y, Ooneda O, et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J Immunol. 2005;175:7348–56. doi: 10.4049/jimmunol.175.11.7348. [DOI] [PubMed] [Google Scholar]
- 112.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu Y, Frith MC, Haverty PM, Weng Z. MotifViz: an analysis and visualization tool for motif discovery. Nucleic Acids Res. 2004;32:W420–3. doi: 10.1093/nar/gkh426. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- 115.Dere E, Boverhof DR, Burgoon LD, Zacharewski TR. In vivo–in vitro toxicogenomic comparison of TCDD-elicited gene expression in Hepa1c1c7 mouse hepatoma cells and C57BL/6 hepatic tissue. BMC Genomics. 2006;7:80. doi: 10.1186/1471-2164-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bjeldanes LF, Kim JY, Grose KR, Bartholemew JC, Bradfield CA. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci USA. 1991;88:9543–7. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–54. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 118.Poland A, Glover E. Comparison of 2,3,7,8-tetrachlorodibenzo-p-dioxin, a potent inducer of aryl hydrocarbon hydroxylase, with 3-methylcholanthrene. Mol Pharmacol. 1974;10:349–59. [PubMed] [Google Scholar]
- 119.Wei YD, Helleberg H, Rannug U, Rannug A. Rapid and transient induction of CYP1A1 gene expression in human cells by the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole. Chem Biol Interact. 1998;110:39–55. doi: 10.1016/s0009-2797(97)00111-7. [DOI] [PubMed] [Google Scholar]
- 120.Spink BC, Hussain MM, Katz BH, Eisele L, Spink DC. Transient induction of cytochromes P450 1A1 and 1B1 in MCF-7 human breast cancer cells by indirubin. Biochem Pharmacol. 2003;66:2313–21. doi: 10.1016/j.bcp.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 121.Schaldach CM, Riby J, Bjeldanes LF. Lipoxin A4: a new class of ligand for the Ah receptor. Biochemistry. 1999;38:7594–600. doi: 10.1021/bi982861e. [DOI] [PubMed] [Google Scholar]
- 122.Sinal CJ, Bend JR. Aryl hydrocarbon receptor-dependent induction of cyp1a1 by bilirubin in mouse hepatoma hepa 1c1c7 cells. Mol Pharmacol. 1997;52:590–9. doi: 10.1124/mol.52.4.590. [DOI] [PubMed] [Google Scholar]
- 123.Song J, Clagett-Dame M, Peterson RE, Hahn ME, Westler WM, Sicinski RR, DeLuca HF. A ligand for the aryl hydrocarbon receptor isolated from lung. Proc Natl Acad Sci USA. 2002;99:14694–9. doi: 10.1073/pnas.232562899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Henry EC, Bemis JC, Henry O, Kende AS, Gasiewicz TA. A potential endogenous ligand for the aryl hydrocarbon receptor has potent agonist activity in vitro and in vivo. Arch Biochem Biophys. 2006;450:67–77. doi: 10.1016/j.abb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 125.Chen YH, Riby J, Srivastava P, Bartholomew J, Denison M, Bjeldanes L. Regulation of CYP1A1 by indolo[3,2-b]carbazole in murine hepatoma cells. J Biol Chem. 1995;270:22548–55. doi: 10.1074/jbc.270.38.22548. [DOI] [PubMed] [Google Scholar]