Figure 2.
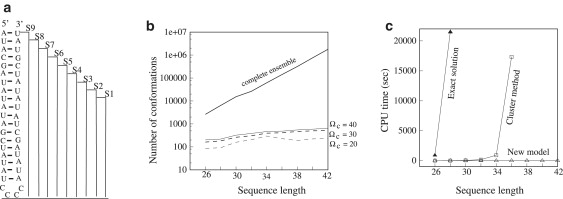
(a) S1–S9 are the nine sequences with different stem lengths and hence different sequence lengths. These sequences are used to compute the data for panel b. (b) The number of conformations from the complete conformational ensemble model and the new kinetic model based on the number of parent states Ωc = 20, 30, and 40. We find the number of conformations from the complete conformational ensemble model grows exponentially as the chain length increases. However, the number of conformations from the new kinetic model increases much more slowly with the chain length. (c) The computational time for the exact master equation (solid triangle), kinetic cluster method (open rectangle), and the new kinetic method (Ωc = 20) (open triangle). The new kinetic method can significantly improve the computational speed. The computational time for the nine sequences is <1 s. In contrast, both the exact master equation and the cluster method are computationally expensive and can only treat short RNA sequences.
