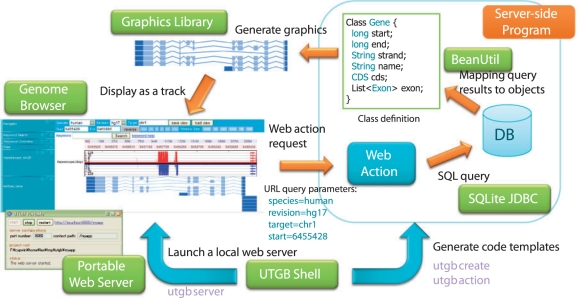
Fig. 3.
Overview of the UTGB Toolkit. The UTGB Toolkit supports development of personalized genome browsers in various ways. The UTGB Shell generates code templates for handling web requests from the genome browser interface (web action), database access support through SQLite JDBC and mapping support from SQL query results to the specified class objects (BeanUtil). Developers can generate track graphics by using the library included in the UTGB Toolkit or their own programs. The generated graphics (or arbitrary HTML contents) can be displayed as track contents in the genome browser interface. To browse both of the locally and remotely stored data, these steps can be performed in a local user machine by launching a local web server from the UTGB Shell.