Figure 3. E1 domain structures and enzymatic mechanism.
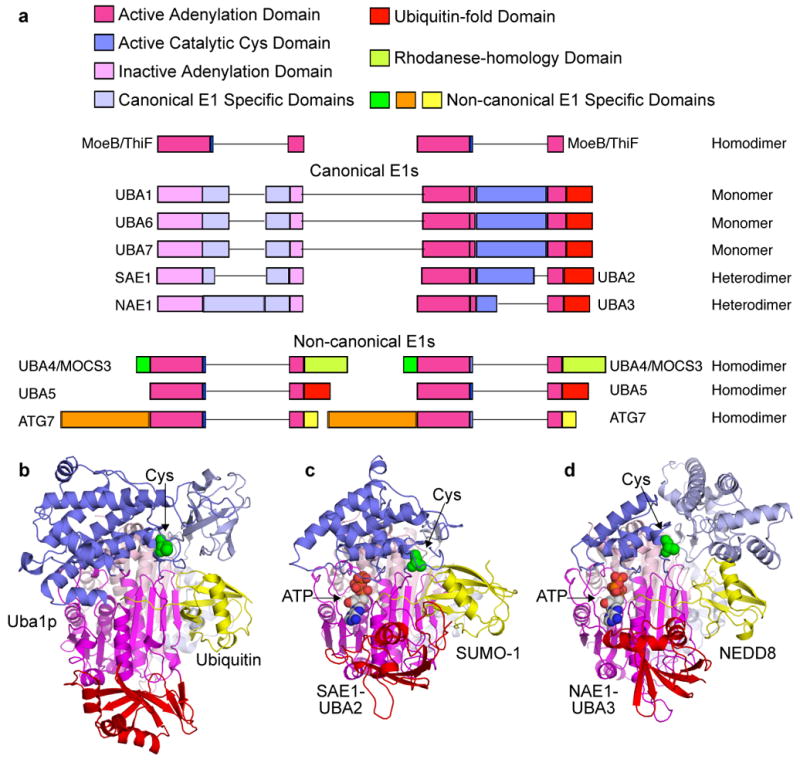
a | Primary structures of human canonical and noncanoncal E1 enzymes, with domains indicated and coloured according to the legend (bottom), and adenylation domains aligned according to molybdopterin biosynthetic enzyme B (MoeB) and thiamine biosynthesis enzyme F (ThiF) primary structures shown above. Lines reflect insertions in sequences between conserved domains. b | Cartoon view of canonical E1 crystal structures with ubiquitin (yellow) associated noncovalently at the adenylation active site of Saccharomyces cerevisiae Uba1 (3CMM.PDB)51, c | with small ubiquitin-related modifier (SUMO)- 1 (yellow) and ATP associated noncovalently at the adenylation active site of SAE1-UBA2 (1Y8R.PDB)50, and d | with NEDD8 (yellow) and ATP noncovalently at the adenylation active site of NAE1-UBA3 (1R4M.PDB)67. The domains are coloured according to the schematic view in panel a, and oriented as the left view of MoeB in FIG. 2a.
