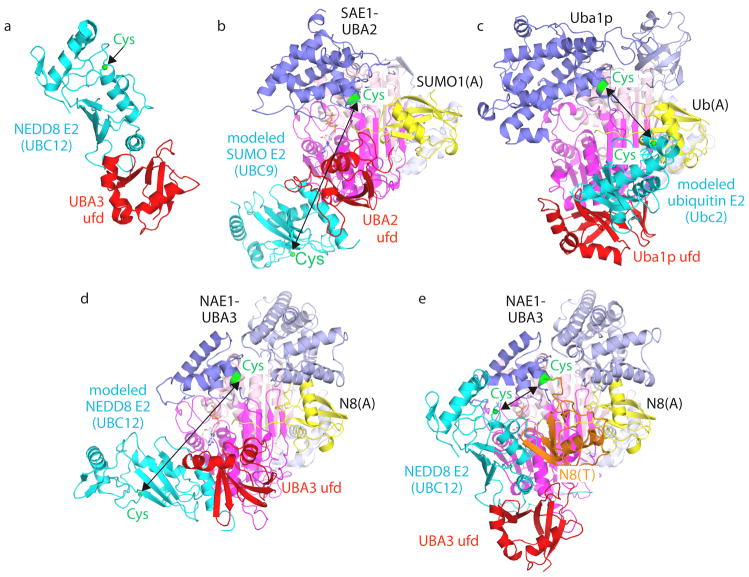
Figure 4. Canonical E1 domain rotation in ubiquitin-like protein transfer to E2s.
a | E1–E2 interactions shown by the crystal structure of the isolated ubiquitin-fold domain (UFD) from the UBA3 subunit of the E1 of NEDD8 (red) complexed with the core domain from UBC12 (cyan), the E2 of NEDD8 (1Y8X.PDB)71. The sulfhydryl of the catalytic Cys in Ubc12 is shown as a green sphere. b–d, Models of E2s (cyan) bound to structures of E1-UBL(A) complexes, with arrows highlighting E1-to-E2 Cys-to-Cys distances, for the E1 of SUMO (1Y8R.PDB)50 and Ubc9 (1U9B.PDB)119 (b), the yeast ubiquitin E1 (3CMM.PDB)51 and Ubc2 (2AYZ.PDB)120 (c), and the E1 of NEDD8 (1R4M.PDB)67 and Ubc12 (1Y8X.PDB)71 (d). e | UFD rotation revealed from the structure of the doubly UBL-loaded/E2-bound NEDD8 E1 [NAE1-UBA3~NEDD8(T)-NEDD8(A)-MgATPUbc12(catalytically inactive mutant] (2NVU.PDB)68. NEDD8(T) is in orange, NEDD8(A) is in yellow, and the location that would correspond to a Ubc12 catalytic Cys is shown as a green sphere. E1-UBL structures are oriented and coloured as in FIG. 3, and E1 and E2 catalytic Cys residues are shown in green.