Abstract
Deposits of amyloid fibrils characterize a diverse group of human diseases that includes Alzheimer disease, Creutzfeldt-Jakob disease and type II diabetes. Amyloid fibrils formed from different polypeptides contain a common cross-β spine. Nevertheless, amyloid fibrils formed from the same polypeptide can occur in a range of structurally different morphologies. The heterogeneity of amyloid fibrils reflects different types of polymorphism: (1) variations in the protofilament number, (2) variations in the protofilament arrangement and (3) different polypeptide conformations. Amyloid fibril polymorphism implies that fibril formation can lead, for the same polypeptide sequence, to many different patterns of inter- or intra-residue interactions. This property differs significantly from native, monomeric protein folding reactions that produce, for one protein sequence, only one ordered conformation and only one set of inter-residue interactions.
Key words: Alzheimer disease, aggregation, neurodegeneration, prion, protein folding
Common Principles of the Amyloid Fibril Architecture
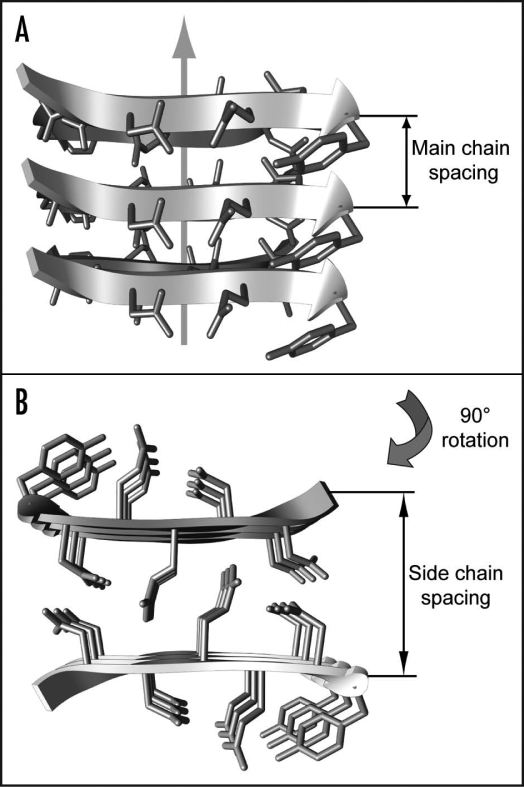
Amyloid fibrils occur inside the human body associated with aging or a group of debilitating health conditions, including type II diabetes, Creutzfeldt-Jakob and Alzheimer diseases.1,2 These fibrils have been defined structurally as fibrillar polypeptide aggregates with a cross-β structure.2 Hence, the central structural spine of these fibrils is formed by an intermolecular and in principle infinite β-sheet structure. The hydrogen bonds that connect juxtaposed β-strands into a pleated β-sheet structure are aligned parallel to the main fibril axis (Fig. 1A). In contrast, the amino acid side chains extend perpendicular to the fibril axis. Therefore, the side chains define the intra- or intermolecular interactions within the plane of the fibril cross-section (Fig. 1B).2
Figure 1.
Cross-β structure of amyloid fibrils. Side (A) and top (B) view of the cross-β core structure as present in microcrystals of peptide fragments of the yeast protein Sup35 (1yjp.pdb).
Presence of a cross-β structure is usually demonstrated by X-ray diffraction analysis. This method reveals a relatively sharp and intense meridional reflection at 4.7 to 4.8 Å, termed main chain spacing, and a more diffuse, equatorial reflection at approximately 10 Å, termed side chain spacing.3 The main chain spacing measures the distance between two hydrogen-bonded β-strands within the same β-sheet (Fig. 1A). The sharpness of this reflection indicates the low variability and high repetition of the underlying molecular unit along the fibril axis. Its conservation for fibrils formed from different polypeptide sequences reflects the fact that all amyloid fibrils encompass the polypeptide main chain in the same basic molecular arrangement.
In contrast, the side chain spacing was shown to vary significantly for different amyloid fibrils.4 Spacings between 8.8 and 14.6 Å were observed. This spacing measures the packing distance between two juxtaposed β-sheets (Fig. 1B). It depends substantially on the sequence of the amino acid side chains that protrude outward from the cross-β sheets. Hence, this variability reflects the differences of the polypeptide sequences constructing the fibril structure. Taken together, X-ray diffraction shows that the structure of amyloid fibrils is highly conserved along the fibril axis, but variable in the plane of the fibril cross-section.
Observation and Biological Relevance of Amyloid Fibril Polymorphism
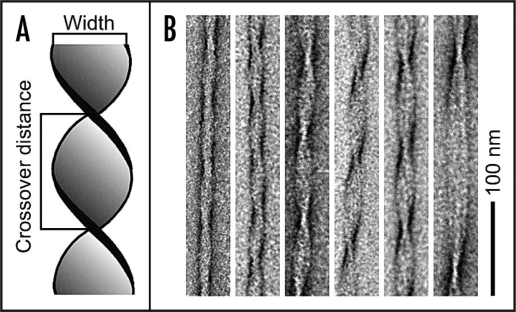
The cross-β sheet structure constructs the core of amyloid protofilaments. These protofilaments represent the filamentous substructures of mature fibrils.5,6 The helical twist of the protofilaments can give rise to a discernible overall helicity of the mature amyloid fibril.7–9 Although the basic structural arrangement of the cross-β structure is conserved for different fibrils, there are different possibilities how they can pack into the three-dimensional fibril structure. Such variable protofilament arrangements can give rise to several distinct amyloid fibril morphologies. Aβ(1–40) peptide, for instance, which forms amyloid fibrils in Alzheimer disease1 produces a broad variety of differently structured amyloid fibrils in vitro.8,10 These fibrils can differ in several structural properties, such as the cross-sectional thickness of the fibril or the helical pitch. Both properties are readily observable by measurements of the fibril width and crossover distance in transmission electron microscopy (TEM) images (Fig. 2). Structural polymorphism of amyloid fibrils has been reported for numerous other polypeptide systems, for example calcitonin,11 amylin,12 glucagon,13 the SH3 domain of phosphatidylinositol-3′-kinase,5,9,13 insulin,6,14,15 lysozyme9 and various Aβ-derived sequences.7,8,16,17
Figure 2.
Polymorphism of Aβ(1–40) fibrils. (A) Schematic representation of an amyloid fibril illustrating the definitions of fibril width and crossover distance. (B) Gallery of negatively stained Aβ(1–40) amyloid fibrils from the same sample.
Structurally polymorphic amyloid fibrils are not only reported for in vitro preparations. Examination of several tissue-extracted amyloid fibrils shows also significant structural polymorphism.18,19 Structurally different fibrils or ensembles of fibril morphologies are thought to underlie different biological activities, such as different toxicities to neuronal cells16,20 or deposition patterns in amyloidotic diseases.21 In several cases the conformational specifics of distinct fibril morphologies were propagated to daughter fibrils by template-dependent seeding.16,22,23 Such self-propagating variations of the molecular fibril structure have been suggested to represent the structural basis of multiple strains of mammalian prion diseases,24,25 different yeast prion phenotypes26,27 and variable clinical or pathological manifestations of several human amyloid disorders.28,29
Definition of Three Structural Types of Amyloid Fibril Polymorphism
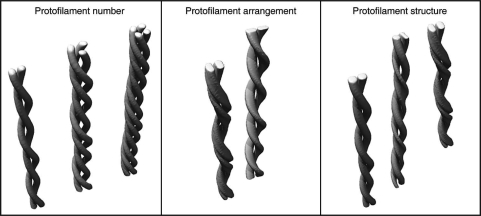
For the purpose of this review, we define three possibilities of how amyloid fibrils can differ in structure (Fig. 3). However, the three types are not mutually exclusive. First, fibrils may consist of a different number of protofilaments. This possibility has been demonstrated by cryo-TEM reconstruction of different insulin fibrils,6 scanning TEM analysis of Aβ(1–40) fibrils8,30 or by atomic force microscopy of amyloid fibrils formed from an immunoglobulin light chain domain.31 Second, fibrils may differ in the relative orientation of their protofilaments. This case was inferred by atomic force microscopy of SH3 domain fibrils9 or cryo-TEM reconstruction of Aβ(1–40) fibrils.10 Third, the fibrils can differ in their protofilament substructure, and therefore, in the conformation of the underlying peptides. Examples hereof were provided by solid-state nuclear magnetic resonance (NMR) spectroscopy or hydrogen/deuterium exchange studies of different fibril morphologies from Aβ(1–40),16 α-synuclein,32 prion protein26,33 and amylin fragments.34 Solid-state NMR studies demonstrated that different Aβ(1–40) amyloid fibril samples can give rise to different NMR spectra.16 For one sample, NMR analysis provided evidence for a single molecular conformation of the Aβ(1–40) peptide, while the other fibril sample evidently contained two structurally different peptide conformations. Moreover, the fibrils in the two samples differ in the precise location of the β-strands along the peptide sequence. While one morphology, termed quiescent fibrils, contains four regions of extended conformation (residues 10–14, 16–22, 30–32 and 34–36), the other morphology, termed agitated fibrils, was reported to encompass continuous β-strand segments at residues 10–22, 30–32 and 34–36. Several other studies also provided evidence for Aβ conformations with variable numbers of β-strands.35–37
Figure 3.
Structural types of amyloid fibril polymorphism. Schematic representation of different amyloid fibril morphologies that differ in the number, relative orientation or structure of the underlying protofilaments.
Molecular Basis of Different Amyloid Fibril Morphologies
The physico-chemical environment substantially influences the fibril morphologies that are stabilized by a given polypeptide chain. The fibril morphology is determined by environmental factors, such as pH value, temperature, agitation, salts or other co-solutes.13,16,26,38–40 Furthermore, certain fibril morphologies can arise by seeding and extension of appropriate structural templates.41 However, even under the same conditions and within the same sample substantial variations of the fibril morphology may exist. Hence, a specific physico-chemical environment does not necessarily lead to a single fibril morphology. In some cases, it rather favors a specific ensemble of fibril structures. This was demonstrated recently for samples of Aβ(1–40) fibrils in the presence of different salts.40 Salts tend to stabilize fibrils with a smaller width, but fibril heterogeneity was observed in fibril samples with and without salts.
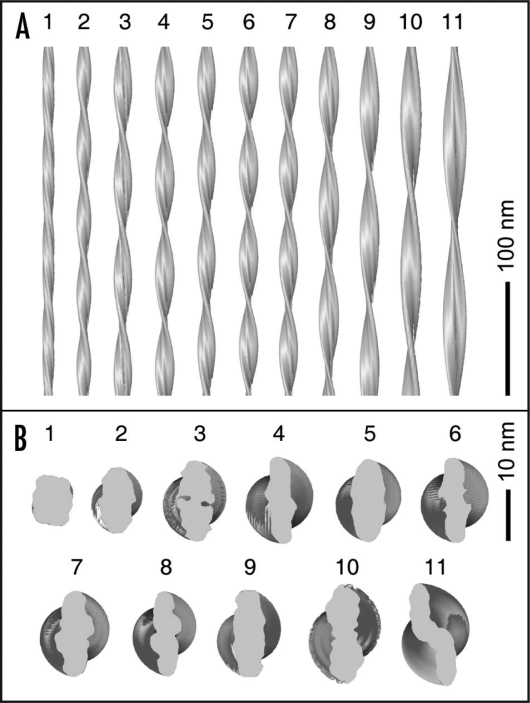
Analysis of the structural diversity of an Aβ(1–40) fibril sample revealed that the individual fibrils could not be classified readily into few, clearly distinct subpopulations.10 Instead, the fibrils formed a morphologic spectrum where structural properties, such as fibril twist or width, vary almost continuously. This result was obtained consistently both by negative stain TEM analysis (Fig. 2) and by structural reconstruction of individual fibrils from cryo-TEM images (Fig. 4). Certain fibril structures, such as fibrils 1, 5 and 11 in Figure 4, possess clearly distinct morphologies. However, these three fibrils represent only certain regions within the entire spectrum of fibril structures, and many intermediate types of fibril structures exist as well. This spectrum of fibrils somewhat resembles the case of an optical spectrum. The latter encompasses also distinctive regions that are commonly termed yellow, red or blue but which are connected by several intermediate colors, altogether constructing the entire spectrum.
Figure 4.
3D densities of Aβ(1–40) amyloid fibrils. Side (A) and top (B) views of cryo-TEM reconstructions of eleven individual Aβ(1–40) fibrils from the same sample. Image reproduced with permission from Meinhardt et al.10
Further studies with other polypeptide systems need to establish the possible general relevance of these observations. However, two main questions arise from the present findings. How can specific fibril morphologies be classified? For example, which of the fibrils shown in Figure 4 belong to the same morphology and why? Unfortunately, answering this question may have to await structural resolution of different fibrils at atomic detail. Second, why is a given polypeptide chain, such as Aβ(1–40), able to adopt so many different fibril structures? The formation of fibril structures with different patterns of interatomic interactions appears to be an intrinsic property of the peptide.
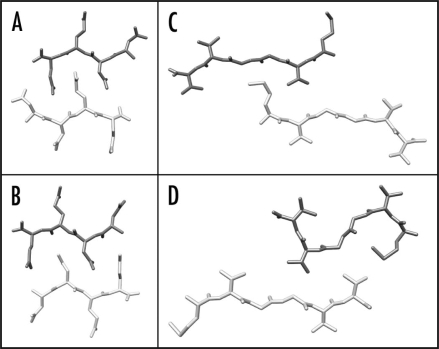
Several studies have shown that the cross-β core structure is formed by only a small proportion of a polypeptide chain, such as Aβ(1–40). The regions forming the β-sheet structure have been identified by various techniques, including peptide fragment analysis, solid-state NMR spectroscopy, hydrogen/deuterium exchange, mutagenesis and proteolysis.35,37,42–44 These studies report slightly different sequence regions of the Aβ(1–40) peptide to be involved in the cross-β core structure. Moreover, the same sequence segments may interact in different ways with each other. This possibility has been demonstrated most clearly for peptide microcrystals that encompass a cross-β structure.45 For example, the peptide fragment NNQQ from Sup35 protein can pack in at least two different manners to form a cross-β structure (Fig. 5A and B). A similar observation was made for microcrystals formed from Aβ(35–40) peptide under different incubation conditions (Fig. 5C and D). Since the structure of amyloid fibrils is highly conserved along the fibril axis (see above) these variations inevitably involve different interactions in the plane of the fibril cross-section and, therefore, different pairings of the amino acid side chains.
Figure 5.
Polymorphism of amyloid-like microcrystals. (A and B) Two different crystal structures of NNQQ peptide, showing either a face-to-back (A, 2onx.pdb) or a face-to-face arrangement (B, 2olx.pdb). (C and D) Two different crystal structures of the Aβ(35–40) fragment, showing different conformations and arrangements of the peptides (2ona.pdb, 2okz.pdb).
The structural heterogeneity and polymorphism of amyloid fibrils represents an important difference from the natively folded protein structure. In native protein folding reactions, a given polypeptide chain always folds up into the same 3D conformation.46,47 Therefore, all natively folded molecules of the same protein sequence share the same inter-residue contacts. Amyloid formation differs from this scenario as the same polypeptide sequence can assume multiple conformationally stable states that are defined by very different inter-residue contacts. These conclusions are in accord with concepts that describe amyloid fibrils as a generic conformational state48,49 or a ‘polymer-state’ of the polypeptide chain.4,50,51 Their nature as organic polymers enables polypeptide chains to form structural states for which sequence specificity is less important than for native protein folding reactions, thus leading to amyloid fibril polymorphism.4,50 More specific explanations of the structural diversity of Aβ(1–40) or other amyloid fibrils, however, need to await structural elucidation of these states at atomic or near-atomic resolutions.
Acknowledgements
The authors thank Carsten Sachse for providing Figure 3. M.F. is supported by grants from BMBF (BioFuture) as well as DFG (SFB 610). J.M. was supported by a grant from the Studienstiftung des deutschen Volkes. N.G. gratefully acknowledges financial support from the National Institutes of Health, grant 1 P01 GM-62580.
Abbreviations
- 3D
three-dimensional
- Aβ
amyloid-β peptide
- NMR, nuclear magnetic resonance
TEM
- transmission electron microscopy
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/8859
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Fändrich M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell Mol Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 4.Fändrich M, Dobson CM. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper JD, Lieber CM, Lansbury PT., Jr Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer's disease amyloid-β protein. Chem Biol. 1997;4:951–959. doi: 10.1016/s1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- 8.Goldsbury CS, Wirtz S, Müller SA, Sunderji S, Wicki P, Aebi U, et al. Studies on the in vitro assembly of Aβ 1–40: implications for the search for Aβ fibril formation inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain AK, MacPhee CE, Zurdo J, Morozova-Roche LA, Hill HA, Dobson CM, et al. Ultrastructural organization of amyloid fibrils by atomic force microscopy. Biophys J. 2000;79:3282–3293. doi: 10.1016/S0006-3495(00)76560-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fändrich M. Aβ(1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol. 2009;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer HH, Aebi U, Häner M, Hermann R, Müller M, Merkle HP. Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J Struct Biol. 1995;115:1–15. doi: 10.1006/jsbi.1995.1024. [DOI] [PubMed] [Google Scholar]
- 12.Goldsbury CS, Cooper GJ, Goldie KN, Müller SA, Saafi EL, Gruijters WTM, et al. Polymorphic fibrillar assembly of human amylin. J Struct Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen JS, Dikov D, Flink JL, Hjuler HA, Christiansen G, Otzen DE. The changing face of glucagon fibrillation: structural polymorphism and conformational imprinting. J Mol Biol. 2006;355:501–523. doi: 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 14.Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzwolak W, Smirnovas V, Jansen R, Winter R. Insulin forms amyloid in a strain-dependent manner: an FT-IR spectroscopic study. Protein Sci. 2004;13:1927–1932. doi: 10.1110/ps.03607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 17.Malinchik SB, Inouye H, Szumowski KE, Kirschner DA. Structural analysis of Alzheimer's β(1–40) amyloid: protofilament assembly of tubular fibrils. Biophys J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez JL, Tennent G, Pepys M, Saibil HR. Structural diversity of ex vivo amyloid fibrils studied by cryo-electron microscopy. J Mol Biol. 2001;311:241–247. doi: 10.1006/jmbi.2001.4863. [DOI] [PubMed] [Google Scholar]
- 19.Crowther RA, Goedert M. Abnormal tau-containing filaments in neurodegenerative diseases. J Struct Biol. 2000;130:271–279. doi: 10.1006/jsbi.2000.4270. [DOI] [PubMed] [Google Scholar]
- 20.Seilheimer B, Bohrmann B, Bondolfi L, Müller F, Stüber D, Döbeli H. The toxicity of the Alzheimer's β-amyloid peptide correlates with a distinct fiber morphology. J Struct Biol. 1997;119:59–71. doi: 10.1006/jsbi.1997.3859. [DOI] [PubMed] [Google Scholar]
- 21.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 22.Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DL. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl Acad Sci USA. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi K, Takahashi S, Kawai T, Naiki H, Goto Y. Seeding-dependent propagation and maturation of amyloid fibril conformation. J Mol Biol. 2005;352:952–960. doi: 10.1016/j.jmb.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 24.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 25.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 26.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 27.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen DL, Hedera P, Premkumar DR, Friedland RP, Kalaria RN. Amyloid-beta protein angiopathies masquerading as Alzheimer's disease? Ann N Y Acad Sci. 1997;826:390–395. doi: 10.1111/j.1749-6632.1997.tb48490.x. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda S, Yanagisawa N. Variable clinical manifestations of familial amyloid polyneuropathy and living related liver transplantation. Rinsho Shinkeigaku. 1995;35:1433–1435. [PubMed] [Google Scholar]
- 30.Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Ionescu-Zanetti C, Khurana R, Gillespie JR, Petrick JS, Trabachino LC, Minert LJ, et al. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:13175–13179. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism and dynamics of full-length a-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Wel PC, Lewandowski JR, Griffin RG. Solid-state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J Am Chem Soc. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 34.Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and X-ray fiber diffraction. J Am Chem Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 35.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, et al. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams AD, Shivaprasad S, Wetzel R. Alanine scanning mutagenesis of Aβ(1–40) amyloid fibril stability. J Mol Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 38.Makarava N, Baskakov IV. The same primary structure of the prion protein yields two distinct self-propagating states. J Biol Chem. 2008;283:15988–15996. doi: 10.1074/jbc.M800562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verel R, Tomka IT, Bertozzi C, Cadalbert R, Kammerer RA, Steinmetz MO, et al. Polymorphism in an amyloid-like fibril-forming model peptide. Angew Chem Int Ed Engl. 2008;47:5842–5845. doi: 10.1002/anie.200800021. [DOI] [PubMed] [Google Scholar]
- 40.Klement K, Wieligmann K, Meinhardt J, Hortschansky P, Richter W, Fändrich M. Effect of different salt ions on the propensity of aggregation and on the structure of Alzheimer's Aβ(1–40) amyloid fibrils. J Mol Biol. 2007;373:1321–1333. doi: 10.1016/j.jmb.2007.08.068. [DOI] [PubMed] [Google Scholar]
- 41.Peim A, Hortschansky P, Christopeit T, Schroeckh V, Richter W, Fändrich M. Mutagenic exploration of the cross-seeding and fibrillation propensity of Alzheimer's β-amyloid peptide variants. Protein Sci. 2006;15:1801–1805. doi: 10.1110/ps.062116206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, et al. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, et al. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of Aβ 1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 45.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 46.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 47.Dobson CM, Sali A, Karplus M. Protein folding: a perspective from theory and experiment. Angew Chem Int Ed. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 48.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fändrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 50.Krebs MR, Macphee CE, Miller AF, Dunlop IE, Dobson CM, Donald AM. The formation of spherulites by amyloid fibrils of bovine insulin. Proc Natl Acad Sci USA. 2004;101:14420–14424. doi: 10.1073/pnas.0405933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wetzel R, Shivaprasad S, Williams AD. Plasticity of amyloid fibrils. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]