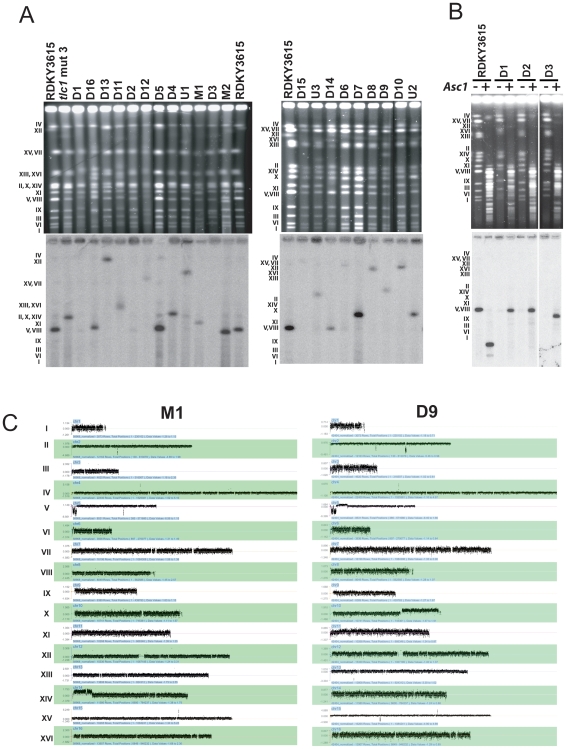
Figure 1. Karyotype analysis of 21 GCRs containing strains by PFGE and aCGH.
(A) PFGE analysis of 21 Canr 5FOAr strains. Intact chromosomes from the indicated GCR strains were transferred to nitrocellulose membranes and hybridized to a radiolabeled chromosome V essential gene YEL058W probe. Rearranged chromosome V sizes were estimated relative to the sizes of chromosomes from the RDKY3615 wild-type and the CANr 5FOAr tlc1 mut 3 strain that were run as controls. (B) Analysis of circular chromosome V GCRs. Circular chromosome V GCRs were digested in the agarose plugs with Asc I for the indicated strains prior to PFGE. The intact and digested chromosome V was detected by hybridization with the YEL058W radiolabelled probe and the size of the resulting chromosome V fragment was estimated. (C) Karyotype analysis by aCGH of representative GCR containing strains is presented. The aGCH data of all GCRs analyzed in this study are present in the Figure S1. The normalized log2 ratio of the fluorescence intensities for each oligonucleotide relative to the reference strain is presented; in order to show all of the data points, it was necessary to use a different scale for the log2 ratio for each chromosome. Chromosome numbers are indicated to the left of the panel.