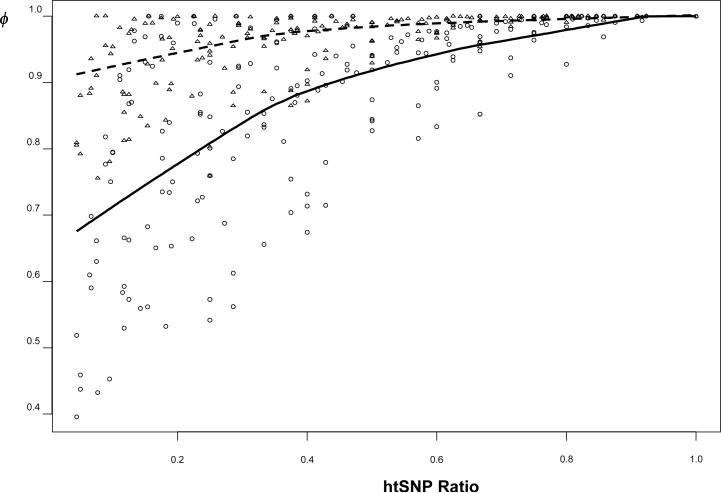
Figure 4.
The relationship between the htSNP ratio (i.e. the size of the htSNP set/the size of the lossless htSNP set) using SMET algorithm and the information capturing ratio defined by either the percentage of coverage based heterozygosity (triangles and the broken line) or based on entropy (circles and the solid line). The datasets used were from SeattleSNPs datasets on NOS3, CRP, IL6, PPARA, the TNF datasets of Ackerman et al. [2003], as well as two region-wide datasets [Daly et al., 2001; Ueda et al. 2003]. The two curves were fit using LOWESS smoothing function with f=2/3. The two curves converge when htSNP ratio = 1 (i.e. when the lossless htSNP set is found).