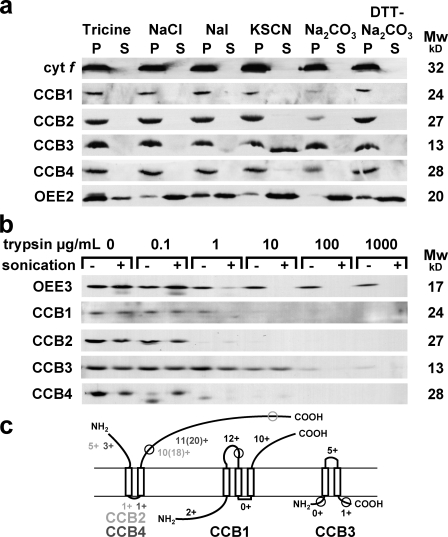
Figure 1.
Transmembrane topology analysis of CCBs. Samples were separated by SDS-PAGE and analyzed by ECL immunodetection with antipeptide antibodies against CCB1, CCB2, CCB3, and CCB4; and with antisera against intrinsic cyt f, extrinsic OEE2, and OEE3 PSII subunits. (a) Extraction of membrane proteins by dissociating treatments. Membranes were frozen and thawed twice in the indicated solutions: 20 mM tricine-NaOH buffer, pH 8.0 (Tricine), 2 M NaCl in 20 mM tricine-NaOH buffer, pH 8.0 (NaCl), 1.5 M NaI in 20 mM tricine-NaOH buffer, pH 8.0 (NaI), 2 M KSCN in 20 mM tricine-NaOH buffer, pH 8.0 (KSCN), 0.1 M Na2CO3 (Na2CO3), and 0.1 M Na2CO3 + 0.1 M DTT (DTT-Na2CO3). The supernatant and the pellet were recovered after a 100,000-g centrifugation. The pellet was extracted a second time using an identical procedure. The first supernatant (S) and the second pellet (P) were analyzed. (b) Proteolysis of membrane vesicles. Membrane vesicles were incubated for 30 min with increasing trypsin protease concentrations (an endoproteinase cutting after positively charged residues in Lys-C and Arg-C) and sonicated (+) or not sonicated (−). (c) Schematized transmembrane topology predicted by sequence analysis and consistent with chaotrope extraction, protease accessibility, and split ubiquitin data. The numbers of positively charged residues after the positive inside rule are indicated for predicted mature protein when a different total number of positively charged residues is also indicated in parenthesis. Antipeptide localizations are circled.