Figure 5. Ubiquitin binds to the peptide-binding hydrophobic groove of the Sla1 SH3-3 domain.
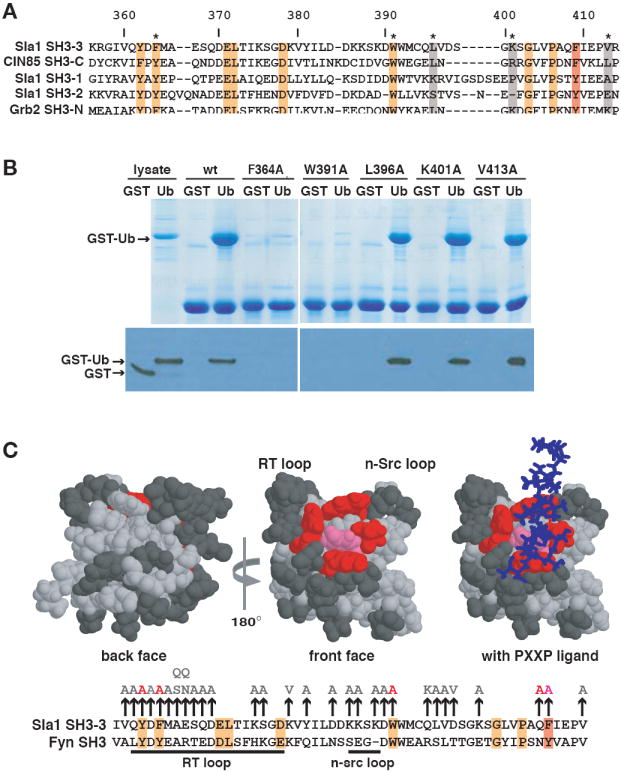
(A) Alignment of Sla1, CIN85 and Grb2 SH3 domains. Numbers above the alignment refer to amino acids of the Sla1 SH3-3 domain. Residues highlighted in orange or red are completely conserved or very similar across the six domains. The position that corresponds to Phe409 of the SH3-3 domain is highlighted with the red box. Non-conserved amino acids that were mutated for the binding analysis shown in (B) are highlighted in grey. (B) The indicated His6-tagged SH3-3 domain mutants immobilized on beads were incubated with lysates containing equivalent amounts of GST or GST-Ub. Lysates or proteins eluted from beads were detected by staining with Coomassie blue (top panel) or on an anti-GST immunoblot (bottom panel). (C) The three-dimensional structures of the Fyn SH3 domain (PDB accession code 1AZG) in the absence and presence of bound PXXP peptide are shown with the residues equivalent to mutated Sla1 SH3-3 domain amino acids highlighted. An alignment of the SH3-3 and Fyn SH3 domain sequences is shown below. Mutation of residues shown in pink (F409) or red (Y362, F364, Q408, W391) abolished ubiquitin-binding. The mutation of residues shown in dark grey had little or no effect on ubiquitin-binding. The position of a PXXP peptide that binds to the Fyn SH3 domain is shown as a stick model in blue. Most of the binding reactions on which these representations are based are shown in (B) or in Supplementary Figure 1. The vertical arrows above the alignment indicate the amino acid substitutions introduced into the SH3-3 domain. Amino acid substitutions in light grey did not affect binding to ubiquitin; the substitutions in red or pink inhibited binding and correspond to residues highlighted on the structural model.
