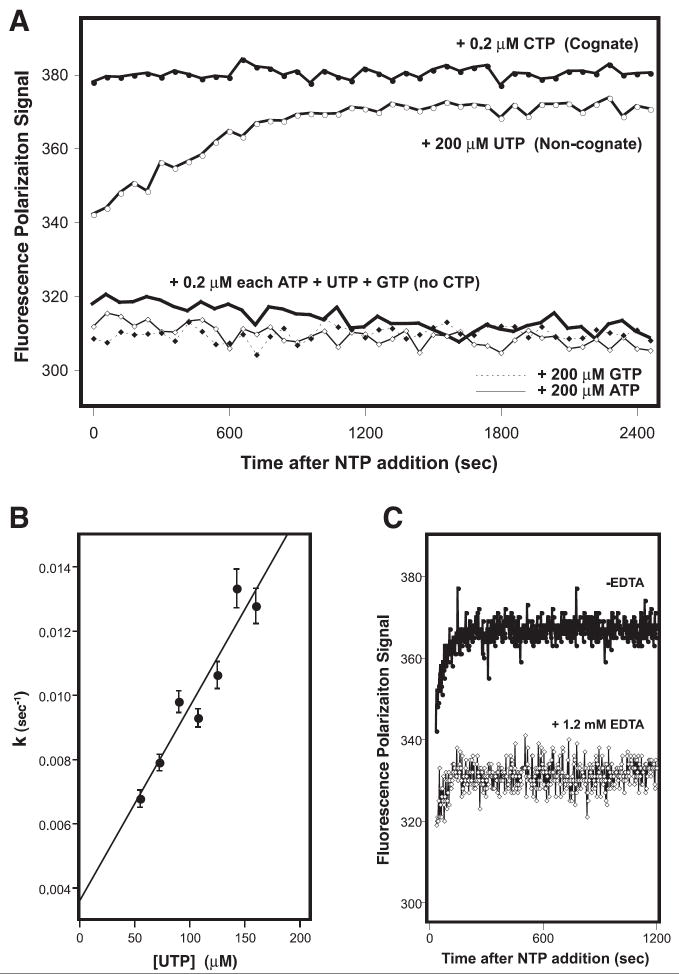
Figure 4.
A) Kinetic analysis of cognate and non-cognate base incorporation by 3Dpol. Polymerase was pre-incubated with the “8-4” RNA hairpin and the 1st, 3rd, and 4th required NTPs at concentrations of 0.2 μM each. This results in a stable complex that has incorporated the first base (A) and is poised for the addition of the missing second nucleotide (C) before fully elongating by incorporating U and G. Addition of the cognate CTP at 0.2 μM concentration results in complete elongation at a rate too rapid to detect by the microplate reader. The incorporation of non-cognate uracil can be driven by increasing the UTP concentration one thousand-fold to 200 μM, resulting in a time dependent exponential increase in the FP signal, while addition of the bulkier ATP or GTP does not drive mis-incorporation. B) Plot showing that the uracil incorporation rate constant is linear with respect to UTP concentration. The rates were obtained by fitting the temporal uracil incorporation curves (e.g. panel A) to a single exponential function. C) Data showing an absence of an increase in the FP signal when the polymerase is inhibited by adding EDTA to chelate the Mg2+ ions required for elongation activity.