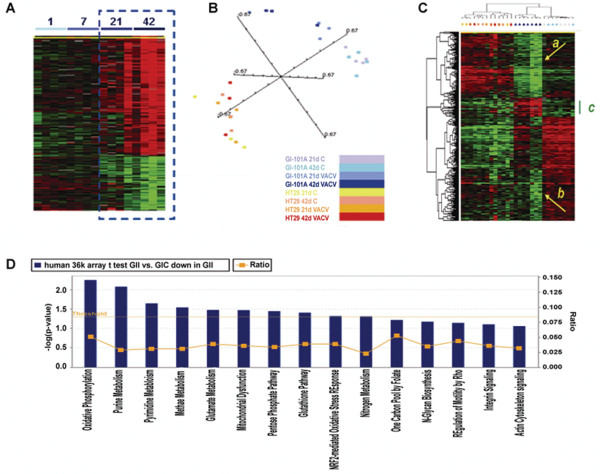
Figure 2.
Human Cancer signatures. (A) Time course analysis of infected GI-101A xenografts (parameters for gene selection; F test p-value < 0.005, 80% presence call, ratio of > 2 and false discovery rate < 0.1). Gene distribution is shown based on 893 genes of 17,500 present in the human cDNA platform that passed the statistical criteria and presented according to Spearman rank correlation. The dashed box outlines the 2 time points most affected by VACV infection. The heat map information is presented according to the central method for normalization [30]. (B) Multiple dimensional scaling based on the 36 k oligo array human platform comparing HT-29 and GI-101 xenografts. (C) Self organizing heat map based on 841 out of 1,073 genes differentially expressed between GI-101A xenografts from infected compared to non-infected mice that passed the standard filter conditions (presence call in at least 80% and at least 3 fold ratio change). HT-29 samples are also represented as a reference, color coding of samples is as per panel (B). The green bar underlines the genes specifically expressed by Gl-101A xenografts from infected animals; the 2 yellow arrows (a) and (b) point at genes who expression was profoundly depressed in xenografts from infected animals. (D) Ingenuity pathway analysis showing canonical pathways significantly down-regulated in GI-101A xenografts at day 41 following GLV-1h68 injection; IPA analysis based on an unpaired, two-tailed Student t test comparing infected to non-infected GI-101A xenografts at day 42 (threshold p2-value < 0.001).