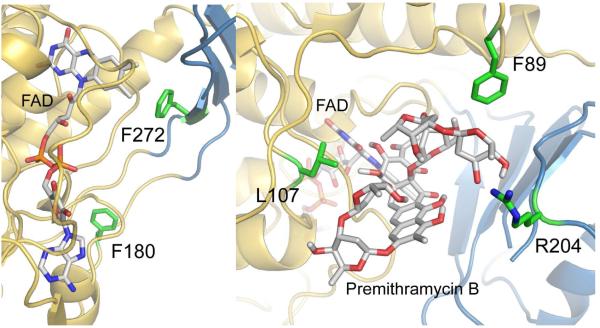
Figure 5.
A. Residues F180 (green), close to the adenine moiety of FAD, and F272 (green), close to the isoalloxazine moiety of FAD (gray stick with N atoms = blue, O-atoms = red and P atoms = orange), were selected for mutagenesis to support the FAD binding area in MtmOIV (gold/blue ribbon). B. Residues L107, R204 and F89 (all in green) were chosen to support their involvement in the substrate (premithramycin B, gray stick with O-atoms = red) binding. The isoalloxazine moiety of FAD (gray stick with N-atoms = blue, O-atoms = red, P-atoms = orange) and the enzyme show in gold/blue ribbon. The results (Table 2) show that residues F89 and R204 possibly constrict the premithramycin B binding site, while the L107A mutation reduces substrate binding, and also FAD binding.