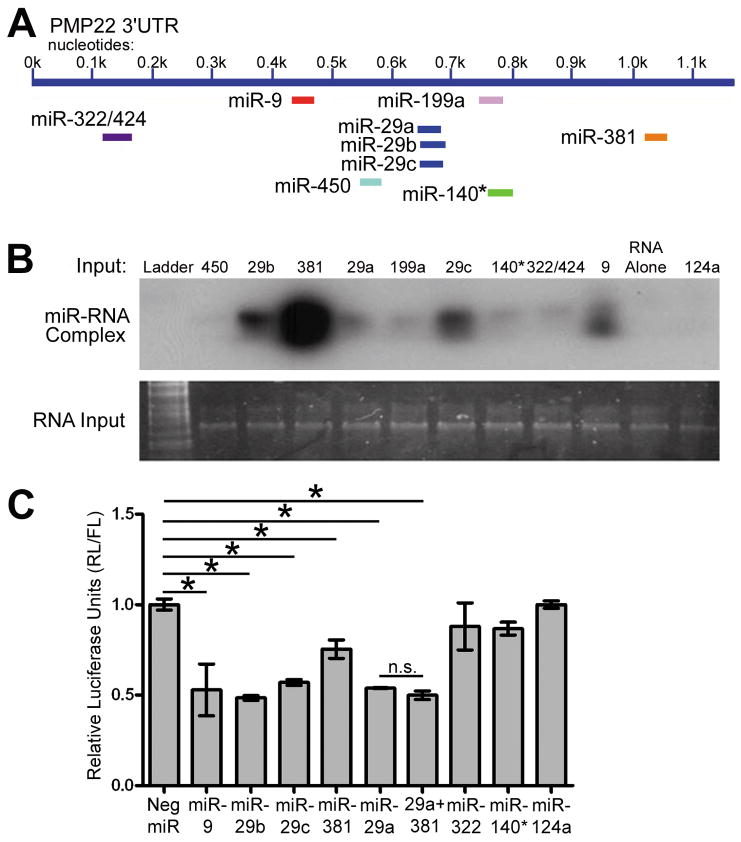
Figure 3. The binding and regulatory ability of predicted PMP22 targeting miRNAs.
(A) MiRNAs that are predicted to bind to the 3′UTR of PMP22 are shown. (B) Binding of candidate miRNAs are detected as miRNA/PMP22 RNA complexes by a gel shift assay. The RNA alone lane contains PMP22 RNA only, whereas miR-124 serves as an additional negative control. Total RNA input is shown using SYBR gold staining. (C) Luciferase assays were performed after co-transfection of the PMP22 3′UTR luciferase reporter and the indicated miRNA-precursors. Luciferase signal is shown after Renilla Luciferase (RL) readings were normalized to Firefly Luciferase (FL). Neg. miR does not code any known miRNA, and miR-124 is used as a non-PMP22 targeting control. (*p<0.05, n=3), n.s.: not significant.