Figure 2. Sequence alignment of gp26-like tail needle homologues.
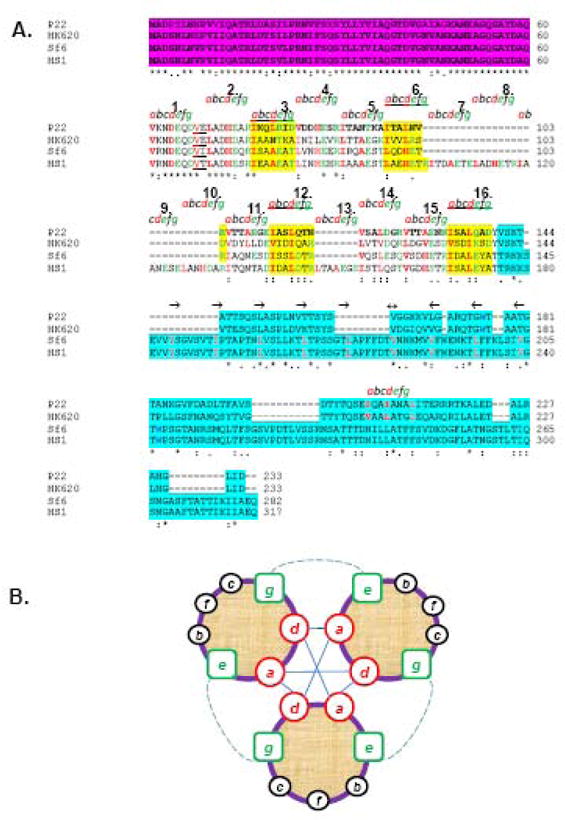
(A) The ClustalW alignment of gp26 homologues from bacteriophage P22, HK620, Sf6 and HS1. Primary sequence of gp26 can be divided into three functional regions, namely, the gp10 binding motif (in magenta), a central helical core containing heptad repeats (labeled as letter a, b, c, d, e, f and g), and the C-terminal domain involved in cell wall penetration (aqua blue). (B) Helical wheel diagram of three-stranded coiled-coils, showing the possible locations of the heptad positions in the central helical core of gp26 structures. Classically, position a and d are occupied by non-polar core residues forming hydrophobic interactions (straight line) whereas e and g are solvent exposed polar residues known to provide specificity between strands through electrostatic interactions (dotted line).
