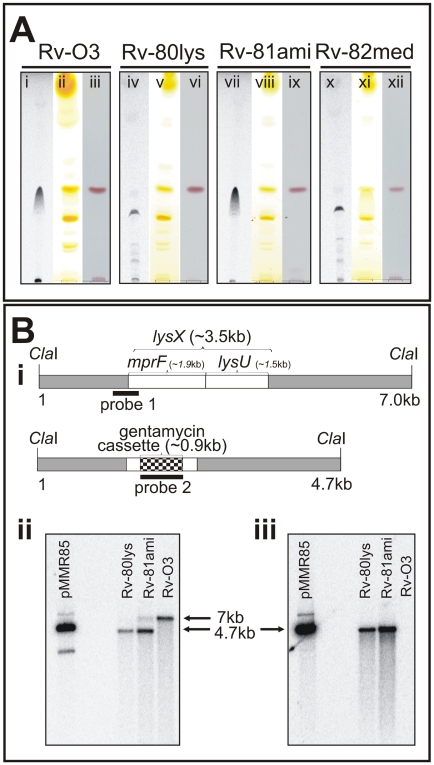
Figure 1. Polar lipid and Southern blot analysis of the lysX mutant strain.
A: Mtb strains were grown in the presence and absence of 14C-lysine. Total lipids were extracted in chloroform∶methanol (2∶1 v/v) and resolved by TLC on Silcia Gel 60 (EMD Chemicals, New Jersey) in a solvent system of chloroform∶methanol∶water (65∶25∶4 v/v/v). TLC plates were either visualized by autoradiography (lanes i, iv, vii and x), exposed to iodine vapors (lanes ii, v, viii and xi), or stained with ninhydrin (lanes iii, vi, ix and xii). B: Southern blot analysis of Mtb lysX mutant strains. B-i: The ClaI fragment bearing the wild type lysX gene (3.5 kb) with the locations of the mprF and lysU regions marked. The dark box designated as “probe 1” is an approximately 750 bp fragment that hybridizes with the 5′-end of lysX and 160 bp of the lysX coding region. The ClaI fragment bearing the mutant lysX gene disrupted with the gentamycin cassette (0.9 kb) is also shown. The dark band designated as “probe 2” is the 900 bp gentamycin gene that hybridizes with the mutant lysX gene. B-ii: Southern blot analysis of ClaI-digested Mtb genomic DNA hybridized with probe 1. The 7 kb and 4 kb band positions represent Rv-03 and Rv-80lys, respectively. Note that the complemented copy contains a band corresponding to the integrated copy of lysX gene plus the flanking plasmid sequence. B-iii: Southern blot analysis of ClaI-digested Mtb genomic DNA (see Fig. 1B-iii) hybridized with probe 2. pMMR85 is a positive control plasmid containing the mutant lysX gene plus flanking regions.