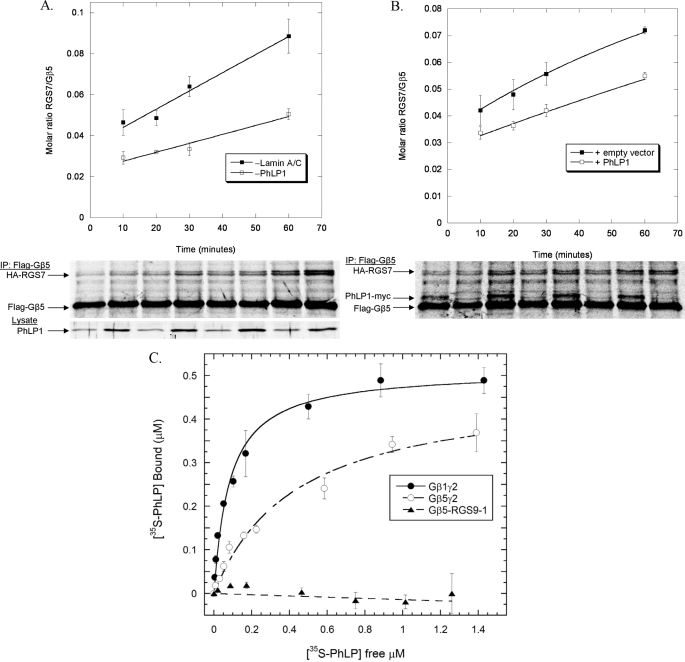
FIGURE 8.
Effects of PhLP1 on the rate of Gβ5-RGS7 dimer formation. A, the rate of Gβ5-RGS7 dimer assembly was measured in HEK-293T cells with or without PhLP1 knockdown. Cells were treated with PhLP1 or lamin A/C siRNAs as indicated. Twenty-four hours later, the cells were transfected with FLAG-Gβ5 and HA-RGS7 cDNAs. After 72 additional h, nascent polypeptides were labeled for 10 min with [35S]methionine and then chased with unlabeled methionine and cycloheximide. At the chase times indicated, the FLAG-Gβ5 was immunoprecipitated and the proteins were separated by SDS-PAGE. The radioactive bands were visualized and quantified using a PhosphorImager, and the molar ratio of Gβ5 to RGS7 was calculated. The data points represent the average ± S.E. from three separate experiments, and lines represent fits of the data to a first order rate equation. A representative gel is shown below the graph as is a PhLP1 immunoblot of 10 μg of whole cell lysate showing the degree of siRNA knockdown. B, HEK-293 cells were transfected with FLAG-Gβ5 and HA-RGS7 with and without PhLP1-Myc cDNAs for 48 h, and the rate of Gβ5-RGS7 assembly was measured using the pulse-chase assay as in panel A. The data are from three separate experiments. C, the binding of the indicated concentrations of 35S-PhLP1 to 0.5 μm purified Gβ1γ2 (○), Gβ5γ2 (○), or Gβ5-RGS9-1 (▴) was measured by in vitro co-immunoprecipitation (see “Experimental Procedures”). Symbols represent the average ± S.E. from three separate experiments. Lines represent non-linear least squares fits of the data to a one-to-one binding equation. The fits yielded Kd values of 83 ± 13 nm for Gβ1γ2, 440 ± 70 nm for Gβ5γ2, and no measurable value for Gβ5-RGS9-1.