Abstract
Purpose
Haploinsufficiency through mutation or deletion of the forkhead transcription factor, FOXC1, causes Axenfeld-Rieger anomaly, which manifests as a range of anterior segment eye defects and glaucoma. The aim of this study is to establish whether mutation of FOXC1 contributes toward other developmental eye anomalies, namely anophthalmia, microphthalmia, and coloboma.
Methods
The coding sequence and 3`-UTR of FOXC1 was analyzed in 114 subjects with severe developmental eye anomalies by bidirectional direct sequencing.
Results
Four coding FOXC1 variations (two novel missense variations, one insertion, and one novel deletion) were identified in the cohort. Two noncoding variations were also identified in the 3′-UTR. The missense mutations were c.889C_T and c.1103C_A, resulting in p.Pro297Ser and p.Thr368Asn, respectively. The c.889C_T transition was identified in 19 of the 100 unaffected control samples. The c.1103C_A transversion resulted in a conservative substitution in an unconserved amino acid and was deemed unlikely to be pathogenic. A c.1142_1144insGCG change resulting in p.Gly380ins, which was previously associated with kidney anomalies, was identified in 44 of the 114 affected individuals. This variation was also present in 29 of the 87 unaffected controls and is therefore likely to be a polymorphism. A c.91_100delCGGCGGCCG deletion resulting in p.Ala31_33del was identified in one individual. This deletion segregated with the moderately affected mother and unaffected maternal grandfather of the proband. This deletion was identified in one of the 307 unaffected controls.
Conclusions
Our data suggests a potential susceptibility role for FOXC1 in generating severe eye pathologies. However, on the basis of these results, it is unlikely that FOXC1 mutation is a major causative factor of anophthalmia, microphthalmia, and coloboma.
Introduction
Developmental eye anomalies (DEA) encompass a spectrum of severe structural defects of the eye caused by the disruption of the smooth process of ocular morphogenesis during early gestation [1]. With a birth prevalence of approximately 1 in 3,000–4,000, DEA are considered to account for at least 25% of childhood visual impairment worldwide [2,3]. The most severe forms of DEA are anophthalmia, characterized by the complete absence of ocular tissue in the orbit, and microphthalmia, which exhibits wide phenotypic variability and causes the eye to have an axial length of two standard deviations below the age-adjusted mean with variable intraocular abnormalities including coloboma [4].
A growing number of monogenic syndromes have begun to be identified in patients exhibiting DEA including those caused by mutations or deletions in orthodenticle homeobox 2 (OTX2), SRY (sex determining region Y)-box 2 (SOX2), H6 family homeobox 1 (NKX5-3), visual system homeobox 2 (CHX10), sonic hedgehog (SHH), retina and anterior neural fold homeobox (RAX), bone morphogenetic protein 4 (BMP4), BCL6 co-repressor (BCOR), chromodomain helicase DNA binding protein 7 (CHD7), and paired box 6 (PAX6) [5-16]. Furthermore, there are up to 400 different chromosome aberrations described in the various dysmorphology databases including deletions, duplications, and translocations [17-23]. Many of these aberrations are likely to act by disrupting eye development genes and likely to harbor additional candidate genes that can be studied to further understand developmental eye disorders. One particular example is the 6p25 deletion syndrome. This syndrome causes deafness [24], developmental delay, facial dysmorphology [25], brachycephaly, schizophrenia [26], and anterior eye anomalies [23-27]. The eye anomalies are thought to be due to a deletion of the forkhead box C1 gene (FOXC1).
FOXC1 is a member of the forkhead family of transcription factors, characterized by their molecular arrangement of two wings connecting β strands flanking one of three α helices [28]. This helix-turn-helix structure comprises the evolutionarily conserved forkhead domain of 110 amino acids through which the FOX proteins are able to interact with DNA and translocate to the cell nuclei [29]. Forkhead genes act as critical regulators of embryogenesis, cell migration, and cell differentiation. Disruptions within the FOX genes have long been associated with pathogenicity and ocular disease in particular [28]. FOXC1 whole gene deletions or mutations within or affecting the forkhead domain through which FOX proteins are able to interact with DNA and translocate to the cell nuclei [29] underlie Axenfeld-Rieger anomalies. To date, at least 30 different missense, nonsense, and frameshift mutations have been identified, affecting the forkhead domain of FOXC1 in individuals presenting with the spectrum of ocular defects associated with Axenfeld-Rieger syndrome and anomaly (anteriorly-displaced Schwalbe’s line, iris adhesions, iridocorneal angle dysgenesis, and corectopia [30-44]). Approximately half of these patients also develop glaucoma, which may cause further visual deterioration. Interestingly, both duplications and deletions of the 6p25 segment containing FOXC1 are associated with anterior eye malformations [25,45]. These seemingly complex genotype-phenotype associations are consistent with FOXC1 gene dosage effects [46]. Intriguingly, one such study by Gould et al. [23] describes seven individuals with 6p25 deletion syndrome associated with ocular dysgenesis of which two individuals presented with microphthalmia.
Since deletions of FOXC1 have been associated with microphthalmia [23], an investigation into the role of FOXC1 in producing developmental eye anomalies, distinct from those associated with Axenfeld-Rieger syndrome, is important in enabling us to delimit the effect of this gene. We therefore decided to investigate a wider role for FOXC1 in underlying developmental eye anomalies and screened the gene for disease-causing variations in a cohort of patients exhibiting anophthalmia, microphthalmia, and coloboma.
Methods
One hundred and fourteen subjects with developmental eye anomalies consisting of unilateral microphthalmia with contralateral normal eye or minor defect such as myopia (n=33); bilateral microphthalmia (n=20) including one with bilateral Peter’s anomaly and one with anterior segment dysgenesis; bilateral anophthalmia (n=12); unilateral anophthalmia with contralateral defect e.g., retinal dystrophy (n=7); unilateral anophthalmia with contralateral coloboma (n=2); unilateral anophthalmia and contralateral microphthalmia (n=3); unilateral anophthalmia with contralateral normal eye or minor defect e.g., myopia (n=11); unilateral coloboma with contralateral normal eye (n=3); unilateral microphthalmia with bilateral coloboma (n=1); unilateral microphthalmia with unilateral coloboma (same eye; n=8); unilateral microphthalmia with contralateral coloboma (n=4); bilateral coloboma (n=2); bilateral microcornea (n=1); unilateral microphthalmia with contralateral defect e.g., retinal dystrophy (n=5); bilateral Peter’s anomaly with normal sized eyes (n=1); and unilateral Peter’s anomaly (n=1) were screened for variations in the coding region of FOXC1 (Ensembl Transcript: FOXC1–001 ENST00000380874). These individuals had been previously screened for mutations in genes known to be associated with anophthalmia, microphthalmia, and coloboma, including SOX2, OTX2, SHH, and BMP4 [5,13]. Informed consent was obtained from all subjects under full ethics approval as previously described [5,13]. Familial DNA and ethnically matched control DNA samples were obtained where possible in the event of a putative causative variant being identified. A control cohort of 100 Yoruba people, 87 CEPH (Centre d'Etude du Polymorphisme Humain) samples, and 307 British Caucasians were analyzed for the variations p. Pro297Ser, p.Gly380ins, and p. Ala31_33del, respectively. Statistical analysis comprising a Fisher's exact test for count data was performed on the data sets using the R software package.
Eight primer pairs spanning the exonic sequence and 3`-UTR of FOXC1 were designed using Primer3 (Table 1) and amplified by polymerase chain reaction (PCR) on a DNA Thermocycler 9700 (Applied Biosystems®, Foster City, CA). PCR was performed according to the manufacturer’s standard protocol in 10 µl reaction volumes using the FailSafeTM PCR System (EPICENTRE® Biotechnologies, Madison, WI) for amplicons FOXC1i, FOXC1ii, and FOXC1iii and Qiagen HotStarTaq DNA Polymerase (Qiagen®, Valencia, CA) for amplicons FOXC1iv, FOXC1v, FOXC1vi, FOXC1vii, and FOXC1viii under the reaction conditions detailed in Table 1.
Table 1. FOXC1 PCR amplifications and primer details.
| Amplicon | Reaction conditions | Annealing temperature | Forward primer sequence | Reverse primer sequence | Fragment length (bp) | Genomic position | Region |
|---|---|---|---|---|---|---|---|
| FOXC1i |
FailSafe – G |
56.5 |
CGGTTCTCACCTCCCATTG |
TTGACGAAGCACTCGTTGAG |
1045 |
chr6: 1555048–1556092 |
Exonic |
| FOXC1ii |
FailSafe – K |
60 |
AGTTCATCATGGACCGCTTC |
ACGTACCGTTCTCGGTCTTG |
381 |
chr6: 1555996–1556376 |
Exonic |
| FOXC1iii |
FailSafe – J |
61 |
GCATCCAGGACATCAAGACC |
CAAGTGGCCCAGGTCTCC |
791 |
chr6: 1556344–1557134 |
Exonic |
| FOXC1iv |
Qiagen |
60 |
CTCACCTCGTGGTACCTGAAC |
AGAGTTTTCTTCGTGCTGGTG |
441 |
chr6: 1557087–1557527 |
Exonic/
3′-UTR |
| FOXC1v |
Qiagen |
60 |
TCCCTCCAAAAATTCAGCTC |
ACGTCAGGTTTTGGGAACAC |
434 |
chr6: 1557482–1557915 |
3′-UTR |
| FOXC1vi |
Qiagen |
60 |
TGGATGTCGTGGACCAAAC |
CTAGCCTCAAAGCAAGCTGAC |
414 |
chr6: 1557869–1558282 |
3′-UTR |
| FOXC1vii |
Qiagen |
59 |
TTTATTTTCCTGCAGCATCTTC |
GATTAAATATCCCTTTCCAACC |
462 |
chr6: 1558222–1558680 |
3′UTR |
| FOXC1viii | Qiagen | 59 | TCCCCCATTTACAATCCTTC | AATCACAGGCCACGTAGAGC | 557 | chr6: 1558597–1559153 | 3′UTR |
Reaction conditions indicate the PCR mix used according to the manufactures standard protocols of the specified kits. The genomic position quoted is the position within Ensembl Transcript: FOXC1-001 ENST00000380874.
Sequencing reactions were performed with the PCR primers on ExoSAP-IT® cleaned products (USB Corporation, Cleveland, OH) using BigDyeTM Terminator (v.3.1) Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Waltham, MA) and resolved on an ABI Prism Genome Analyzer 3700 or 3100 for longer reads (Applied Biosystems®). Data were analyzed by constructing contigs aligned to a reference sequence using SequencherTM software (v.4.5; Gene Codes Corporation, Ann Arbor, MI). Confirmation of the sequence changes was obtained using a second sample.
Results
Four FOXC1 coding sequence alterations were revealed by direct sequencing comprising two missense variations, one insertion, and one deletion. Two noncoding variations were also identified (Table 2 and Figure 1).
Table 2. Detected variations and disease phenotype.
| Variation | Phenotype |
|---|---|
| Heterozygous p.Pro297Ser |
Unilateral microphthalmia and sclerocornea |
| Heterozygous p.Pro297Ser |
Unilateral extreme microphthalmia with cyst, contralateral myopia |
| Heterozygous p.Thr368Asn |
Unilateral microphthalmia and dense cataract |
| Heterozygous p.Gly380ins |
Multiple patients |
| Homozygous p.Gly380ins |
Multiple patients |
| Heterozygous p.Ala31_33del |
Right optic disc coloboma and left iris and chorioretinal coloboma |
| Heterozygous 3`-UTR 1662+1041C>T |
Right optic disc coloboma and left iris and chorioretinal coloboma |
| Heterozygous 3`-UTR 1622+396delC |
Bilateral microphthalmia with Rieger anomaly and |
| Heterozygous 3`-UTR 1622+396delC | Unilateral microphthalmia with microcornea and subtotal retinal detachment |
All variations detected within the patient cohort with their corresponding phenotype.
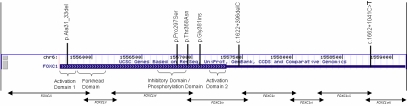
Figure 1.
FOXC1 gene structure. Locations of amplicons and identified variations are displayed.
The deletion (c.91_100delCGGCGGCCG; Figure 2D) caused a three residue contraction of the alanine tract (p.Ala31_33del), located 41 amino acids upstream of the forkhead domain in activation domain 1 of the protein [29]. The deletion resulted in the removal of a SacII recognition site from the PCR product. This deletion was identified in one individual who presented with an optic disc coloboma in both eyes with additional iris and chorioretinal coloboma in the left eye. The moderately affected mother and unaffected maternal grandfather were all heterozygous for the deletion. Three hundred and seven ethnically matched control DNA samples of British Caucasian origin and the family members of the proband (parents, maternal grandparents, sister) were screened for the variation using a restriction digest method exploiting the absence of the SacII restriction site. One control sample was also heterozygous for c.91_100delCGGCGGCCG. These frequencies were analyzed using Fisher’s exact test and found not to be significant (p=0.4678)
Figure 2.
Direct sequencing analysis of the coding region of FOXC1. A: The lower sequence shows the heterozygous missense mutation c889C_T (p.Pro297Ser) identified in two individuals, and the upper sequence is the corresponding wild-type sequence. B: The lower sequence shows the heterozygous missense mutation c.1103C_A (p.Thr368Asn) detected in one individual, and the upper sequence is the corresponding wild-type sequence. C: The bottom sequence shows the 3-bp heterozygous insertion mutation, c.1142_1144insGCG (p.Gly380ins), detected in 37 individuals. The middle sequence shows the equivalent homozygous insertion detected in seven individuals. The upper sequence displays the corresponding wild-type sequence. D: The lower sequence shows the 9-bp heterozygous deletion, c.91_100delCGGCGGCCG (p.Ala31_33del), detected in the proband, mother, maternal grandfather, and 1 of the 307 control samples, and the upper sequence displays the corresponding wild-type sequence.
The first missense mutation was present in two patients, one of Nigerian and the other of Hispanic descent. It resulted from a c.889C_T transition (Figure 2A), causing a p.Pro297Ser amino acid alteration in the protein inhibitory/phosphorylation domain (as defined by Berry et al. [29]). This variation resulted in the introduction of an HgaI restriction site into the gene. One hundred ethnically matched DNA samples of Yoruba descent were screened for the variation in a restriction digest assay with HgaI. The variation was identified in 19 of the control samples.
The second missense mutation, c.1103C_A (Figure 2B), resulted in p.Thr368Asn outside any of the defined functional domains of the protein. This variation was present in a patient of Singaporean-Filipino descent and resulted in a conservative amino acid substitution in a residue, which was highly variable across species. No familial or ethnically matched control DNA sample was available for this variant and thus could not be investigated further. SIFT (Sorting Intolerant From Tolerant) analysis [47] supported the low likelihood of p.Thr368Asn being a pathogenic variant.
The final variation identified was a c.1142_1144insGCG change (Figure 2C), resulting in p.Gly381ins, which was present in a heterozygous form in 37 individuals and a homozygous form in seven individuals. This variation has been previously reported in a screen of patients with congenital abnormalities of the kidney and urinary tract [48]. The heterozygous form of this variation was identified in 27 out of 87 unaffected control samples and was present as a homozygous variant in two of the control samples. Fisher’s exact test analysis of these frequencies indicated that they were not significant (p=0.4523)
Discussion
We report the first screen of patients manifesting with severe developmental eye anomalies for disease-causing variations in FOXC1. Since deletions of FOXC1 have been associated with microphthalmia [23], an investigation into the role of FOXC1 in producing developmental eye defects distinct from those previously associated with the mutation in this gene is important to enable us to delimit its effect. To date, mutations of human FOXC1 have been associated with anterior segment dysgenesis, iris anomalies, and developmental glaucoma. These phenotypes may arise from an abnormality in the migration and/or differentiation of mesenchymal cells that contribute to the anterior segment of the eye as in the mouse. Kume et al. [49] demonstrated that 16.5 dpc (days post coitum) Foxc1 null mice display several ocular anomalies including a disorganized arrangement of cells in the cornea, iris hypoplasia, unfused eyelids, and a reduced number of mesenchymal cells in the future stromal region.
Although an important role for FOXC1 in eye development is clear, the complexities of this association are evident through the lack of any conclusive genotype-phenotype correlations; the eye phenotypes associated with 6p25 deletion syndrome often exhibit variable penetrance [25]. Individuals with FOXC1 mutations or deletions also demonstrate a spectrum of phenotypic consequences, including the mutated allele segregating with affected and unaffected members of the same family [36,39-41]. It has therefore been suggested that either environmental factors and/or modifier genes interact with FOXC1 in producing a disease phenotype [50], and this is not uncommonly seen with other ocular developmental genes [13].
The level of phenotypic variability could also be attributed to stochastic factors in development related to spatio-temporal events and the level of expression of developmentally important downstream targets of FOXC1 [50]. Recent studies have identified one of those downstream targets as another forkhead transcription factor, FOXO1A [51]. The zebrafish foxO1a ortholog is strongly expressed in the periocular mesenchyme, and its expression pattern is significantly reduced in a foxc1 siRNA knocked down model. The reduced foxc1 expression increases cell death in the developing zebrafish eye and demonstrates a novel role for FOXC1 as an essential mediator of cellular homeostasis in the eye.
Unaffected control DNA analysis of c.889C_T demonstrated a high likelihood of this variation being a common polymorphism in the Yoruba population with the variant segregating with 19 out of 100 control samples. The second missense mutation, c.1103C_A, was not analyzed further due to the low likelihood of it being a pathogenic variant.
The c.1142_1144insGCG insertion had been previously reported in a study of patients with congenital abnormalities of the kidney and urinary tract [48]. This variation was present in three of the seven patients presented in the study and was postulated to be causative for the disease phenotype. The identification of this variation in 29 of the 87 controls screened in our study suggests that this variation is a non-pathogenic polymorphism.
The most interesting variation identified in our screen was the deletion resulting in the contraction of the alanine tract to three residues by the deletion. The alanine tract is originally already very short, consisting of only six residues. A polyalanine tract upstream of the forkhead domain is a feature common to all of the forkhead transcription factors and variations in the length of polyalanine repeats have previously been demonstrated to underlie disease phenotypes, e.g., FOXL2 in blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) [52] and FOXE1 in thyroid dysgenesis [53]. In the FOXE1/thyroid dysgenesis model, there is a significant association between a shorter polyalanine tract and the manifestation of the disease phenotype. As a disease-causing mechanism, Carré et al. [53] demonstrate that the transcriptional activation of FOXE1 with 16 alanines is significantly higher than FOXE1 containing 14 alanines. They concluded that FOXE1 significantly modulates the risk of thyroid dysgenesis occurrence through its alanine-containing stretch and proposed a mechanism linking the polyalanine tract containing transcription factors to disease. Interestingly, the disease-associated variant in FOXE1 also segregates with unaffected controls, which is similar to our observation with the contraction of the polyalanine tract in FOXC1. Therefore, this is consistent with the contraction of alanine tract being a susceptibility factor rather than a disease-causing mutation.
The oligogenic basis of developmental eye anomalies is well recognized and will only be resolved when a comprehensive candidate gene set has been analyzed for mutations, coding polymorphisms, or copy number variations. In this study, we have identified several variations affecting the coding sequence of FOXC1, some of which could contribute to phenotype severity and penetrance. Although a direct causative role for FOXC1 mutations in our cohort of patients with developmental eye anomalies has not been definitively shown, FOXC1 could contribute genetic susceptibility either through contraction in the length of the polyalanine tract or other genetic variations similar to the ones described here.
Acknowledgments
The authors would like to thank Laura Winchester for her assistance with the data analysis. This work was funded by the Wellcome Trust. NR is funded by an Academic of Medical Sciences/Health Foundation Senior Surgical Scientist Award.
References
- 1.Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr Opin Genet Dev. 2005;15:348–53. doi: 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Hornby SJ, Gilbert CE, Rahi JK, Sil AK, Xiao Y, Dandona L, Foster A. Regional variation in blindness in children due to microphthalmos, anophthalmos and coloboma. Ophthalmic Epidemiol. 2000;7:127–38. [PubMed] [Google Scholar]
- 3.Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakrania P, Efthymiou M, Klein JC, Salt A, Bunyan DJ, Wyatt A, Ponting CP, Martin A, Williams S, Lindley V, Gilmore J, Restori M, Robson AG, Neveu MM, Holder GE, Collin JR, Robinson DO, Farndon P, Johansen-Berg H, Gerrelli D, Ragge NK. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am J Hum Genet. 2008;82:304–19. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakrania P, Robinson DO, Bunyan DJ, Salt A, Martin A, Crolla JA, Wyatt A, Fielder A, Ainsworth J, Moore A, Read S, Uddin J, Laws D, Pascuel-Salcedo D, Ayuso C, Allen L, Collin JR, Ragge NK. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br J Ophthalmol. 2007;91:1471–6. doi: 10.1136/bjo.2007.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferda Percin E, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AM, Kalnins VI, Kocak-Altintas A, Sowden JC, Traboulsi E, Sarfarazi M, McInnes RR. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 8.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 9.Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, Clarke MP, Russell-Eggitt I, Fielder A, Gerrelli D, Martinez-Barbera JP, Ruddle P, Hurst J, Collin JR, Salt A, Cooper ST, Thompson PJ, Sisodiya SM, Williamson KA, Fitzpatrick DR, van Heyningen V, Hanson IM. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–22. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, de Sanctis U, Salt A, Collin JR, Vivian AJ, Free SL, Thompson P, Williamson KA, Sisodiya SM, van Heyningen V, Fitzpatrick DR. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. discussion 8. [DOI] [PubMed] [Google Scholar]
- 11.Schimmenti LA, de la Cruz J, Lewis RA, Karkera JD, Manligas GS, Roessler E, Muenke M. Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. Am J Med Genet A. 2003;116A:215–21. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- 12.Voronina VA, Kozhemyakina EA, O’Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–22. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 13.Wyatt A, Bakrania P, Bunyan DJ, Osborne Crolla JA, Salt A, Ayuso C, Newbury-Ecob R, Abou-Rayyah Y, Collin JR, Robinson D, Ragge N. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum Mutat. 2008;29:E278–83. doi: 10.1002/humu.20869. [DOI] [PubMed] [Google Scholar]
- 14.Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, van der Smagt JJ, Gorlin RJ, Burgess SM, Bardwell VJ, Black GC, Biesecker LG. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–6. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 15.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–7. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 16.Schorderet DF, Nichini O, Boisset G, Polok B, Tiab L, Mayeur H, Raji B, de la Houssaye G, Abitbol MM, Munier FL. Mutation in the human homeobox gene NKX5–3 causes an oculo-auricular syndrome. Am J Hum Genet. 2008;82:1178–84. doi: 10.1016/j.ajhg.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolen LD, Amor D, Haywood A, St Heaps L, Willcock C, Mihelec M, Tam P, Billson F, Grigg J, Peters G, Jamieson RV. Deletion at 14q22–23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140:1711–8. doi: 10.1002/ajmg.a.31335. [DOI] [PubMed] [Google Scholar]
- 18.Male A, Davies A, Bergbaum A, Keeling J, FitzPatrick D, Mackie Ogilvie C, Berg J. Delineation of an estimated 6.7 MB candidate interval for an anophthalmia gene at 3q26.33-q28 and description of the syndrome associated with visible chromosome deletions of this region. Eur J Hum Genet. 2002;10:807–12. doi: 10.1038/sj.ejhg.5200890. [DOI] [PubMed] [Google Scholar]
- 19.Jamieson RV, Gaunt L, Donnai D, Black GC, Kerr B, Stecko O, Black GC. Chromosomal translocation in a family with ocular anomalies: indications for karyotype analysis. Br J Ophthalmol. 2003;87:646–8. doi: 10.1136/bjo.87.5.646-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–3. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 21.Driggers RW, Macri CJ, Greenwald J, Carpenter D, Avallone J, Howard-Peebles PN, Levin SW. Isolated bilateral anophthalmia in a girl with an apparently balanced de novo translocation: 46,XX,t(3;11)(q27;p11.2). Am J Med Genet. 1999;87:201–2. doi: 10.1002/(sici)1096-8628(19991126)87:3<201::aid-ajmg1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.de Azevedo Moreira LM, Neri FB, de Quadros Uzeda S, de Carvalho AF, Santana GC, Souza FR, Rollemberg JC. Multiple congenital malformations including severe eye anomalies and abnormal cerebellar development with Dandy-Walker malformation in a girl with partial trisomy 3q. Ophthalmic Genet. 2005;26:37–43. doi: 10.1080/13816810590927217. [DOI] [PubMed] [Google Scholar]
- 23.Gould DB, Jaafar MS, Addison MK, Munier F, Ritch R, MacDonald IM, Walter MA. Phenotypic and molecular assessment of seven patients with 6p25 deletion syndrome: relevance to ocular dysgenesis and hearing impairment. BMC Med Genet. 2004;5:17. doi: 10.1186/1471-2350-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies AF, Mirza G, Sekhon G, Turnpenny P, Leroy F, Speleman F, Law C, van Regemorter N, Vamos E, Flinter F, Ragoussis J. Delineation of two distinct 6p deletion syndromes. Hum Genet. 1999;104:64–72. doi: 10.1007/s004390050911. [DOI] [PubMed] [Google Scholar]
- 25.Mirza G, Williams RR, Mohammed S, Clark R, Newbury-Ecob R, Baldinger S, Flinter F, Ragoussis J. Refined genotype-phenotype correlations in cases of chromosome 6p deletion syndromes. Eur J Hum Genet. 2004;12:718–28. doi: 10.1038/sj.ejhg.5201194. [DOI] [PubMed] [Google Scholar]
- 26.Caluseriu O, Mirza G, Ragoussis J, Chow EW, MacCrimmon D, Bassett AS. Schizophrenia in an adult with 6p25 deletion syndrome. Am J Med Genet A. 2006;140:1208–13. doi: 10.1002/ajmg.a.31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann OJ, Ebenezer ND, Ekong R, Ocaka L, Mungall AJ, Fraser S, McGill JI, Hitchings RA, Khaw PT, Sowden JC, Povey S, Walter MA, Bhattacharya SS, Jordan T. Ocular developmental abnormalities and glaucoma associated with interstitial 6p25 duplications and deletions. Invest Ophthalmol Vis Sci. 2002;43:1843–9. [PubMed] [Google Scholar]
- 28.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox's in development and disease. Trends Genet. 2003;19:339–44. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 29.Berry FB, Saleem RA, Walter MA. FOXC1 transcriptional regulation is mediated by N- and C-terminal activation domains and contains a phosphorylated transcriptional inhibitory domain. J Biol Chem. 2002;277:10292–7. doi: 10.1074/jbc.M110266200. [DOI] [PubMed] [Google Scholar]
- 30.Weisschuh N, Wolf C, Wissinger B, Gramer E. A novel mutation in the FOXC1 gene in a family with Axenfeld-Rieger syndrome and Peters' anomaly. Clin Genet. 2008;74:476–80. doi: 10.1111/j.1399-0004.2008.01025.x. [DOI] [PubMed] [Google Scholar]
- 31.Weisschuh N, Dressler P, Schuettauf F, Wolf C, Wissinger B, Gramer E. Novel mutations of FOXC1 and PITX2 in patients with Axenfeld-Rieger malformations. Invest Ophthalmol Vis Sci. 2006;47:3846–52. doi: 10.1167/iovs.06-0343. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Takahashi K, Kuwahara S, Wada Y, Abe T, Tamai M. A novel (Pro79Thr) mutation in the FKHL7 gene in a Japanese family with Axenfeld-Rieger syndrome. Am J Ophthalmol. 2001;132:572–5. doi: 10.1016/s0002-9394(01)01059-5. [DOI] [PubMed] [Google Scholar]
- 33.Saleem RA, Murphy TC, Liebmann JM, Walter MA. Identification and analysis of a novel mutation in the FOXC1 forkhead domain. Invest Ophthalmol Vis Sci. 2003;44:4608–12. doi: 10.1167/iovs.03-0090. [DOI] [PubMed] [Google Scholar]
- 34.Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA. Structural and functional analyses of disease-causing missense mutations in the forkhead domain of FOXC1. Hum Mol Genet. 2003;12:2993–3005. doi: 10.1093/hmg/ddg324. [DOI] [PubMed] [Google Scholar]
- 35.Panicker SG, Sampath S, Mandal AK, Reddy AB, Ahmed N, Hasnain SE. Novel mutation in FOXC1 wing region causing Axenfeld-Rieger anomaly. Invest Ophthalmol Vis Sci. 2002;43:3613–6. [PubMed] [Google Scholar]
- 36.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–7. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 37.Mortemousque B, Amati-Bonneau P, Couture F, Graffan R, Dubois S, Colin J, Bonneau D, Morissette J, Lacombe D, Raymond V. Axenfeld-Rieger anomaly: a novel mutation in the forkhead box C1 (FOXC1) gene in a 4-generation family. Arch Ophthalmol. 2004;122:1527–33. doi: 10.1001/archopht.122.10.1527. [DOI] [PubMed] [Google Scholar]
- 38.Mirzayans F, Gould DB, Héon E, Billingsley GD, Cheung JC, Mears AJ, Walter MA. Axenfeld-Rieger syndrome resulting from mutation of the FKHL7 gene on chromosome 6p25. Eur J Hum Genet. 2000;8:71–4. doi: 10.1038/sj.ejhg.5200354. [DOI] [PubMed] [Google Scholar]
- 39.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerbäck S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–28. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatireddy S, Chakrabarti S, Mandal AK, Reddy AB, Sampath S, Panicker SG, Balasubramanian D. Mutation spectrum of FOXC1 and clinical genetic heterogeneity of Axenfeld-Rieger anomaly in India. Mol Vis. 2003;9:43–8. [PubMed] [Google Scholar]
- 41.Khan AO, Aldahmesh MA, Al-Amri A. Heterozygous FOXC1 mutation (M161K) associated with congenital glaucoma and aniridia in an infant and a milder phenotype in her mother. Ophthalmic Genet. 2008;29:67–71. doi: 10.1080/13816810801908152. [DOI] [PubMed] [Google Scholar]
- 42.Ito YA, Footz TK, Murphy TC, Courtens W, Walter MA. Analyses of a novel L130F missense mutation in FOXC1. Arch Ophthalmol. 2007;125:128–35. doi: 10.1001/archopht.125.1.128. [DOI] [PubMed] [Google Scholar]
- 43.Honkanen RA, Nishimura DY, Swiderski RE, Bennett SR, Hong S, Kwon YH, Stone EM, Sheffield VC, Alward WL. A family with Axenfeld-Rieger syndrome and Peters Anomaly caused by a point mutation (Phe112Ser) in the FOXC1 gene. Am J Ophthalmol. 2003;135:368–75. doi: 10.1016/s0002-9394(02)02061-5. [DOI] [PubMed] [Google Scholar]
- 44.Fuse N, Takahashi K, Yokokura S, Nishida K. Novel mutations in the FOXC1 gene in Japanese patients with Axenfeld-Rieger syndrome. Mol Vis. 2007;13:1005–9. [PMC free article] [PubMed] [Google Scholar]
- 45.Ekong R, Jeremiah S, Judah D, Lehmann O, Mirzayans F, Hung YC, Walter MA, Bhattacharya S, Gant TW, Povey S, Wolfe J. Chromosomal anomalies on 6p25 in iris hypoplasia and Axenfeld-Rieger syndrome patients defined on a purpose-built genomic microarray. Hum Mutat. 2004;24:76–85. doi: 10.1002/humu.20059. [DOI] [PubMed] [Google Scholar]
- 46.Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, Sheffield VC. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet. 2001;68:364–72. doi: 10.1086/318183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakano T, Niimura F, Hohenfellner K, Miyakita E, Ichikawa I. Screening for mutations in BMP4 and FOXC1 genes in congenital anomalies of the kidney and urinary tract in humans. Tokai J Exp Clin Med. 2003;28:121–6. [PubMed] [Google Scholar]
- 49.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–96. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 50.Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA. Analyses of the effects that disease-causing missense mutations have on the structure and function of the winged-helix protein FOXC1. Am J Hum Genet. 2001;68:627–41. doi: 10.1086/318792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry FB, Skarie JM, Mirzayans F, Fortin Y, Hudson TJ, Raymond V, Link BA, Walter MA. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum Mol Genet. 2008;17:490–505. doi: 10.1093/hmg/ddm326. [DOI] [PubMed] [Google Scholar]
- 52.Harris SE, Chand AL, Winship IM, Gersak K, Aittomäki K, Shelling AN. Identification of novel mutations in FOXL2 associated with premature ovarian failure. Mol Hum Reprod. 2002;8:729–33. doi: 10.1093/molehr/8.8.729. [DOI] [PubMed] [Google Scholar]
- 53.Carré A, Castanet M, Sura-Trueba S, Szinnai G, Van Vliet G, Trochet D, Amiel J, Léger J, Czernichow P, Scotet V, Polak M. Polymorphic length of FOXE1 alanine stretch: evidence for genetic susceptibility to thyroid dysgenesis. Hum Genet. 2007;122:467–76. doi: 10.1007/s00439-007-0420-5. [DOI] [PubMed] [Google Scholar]