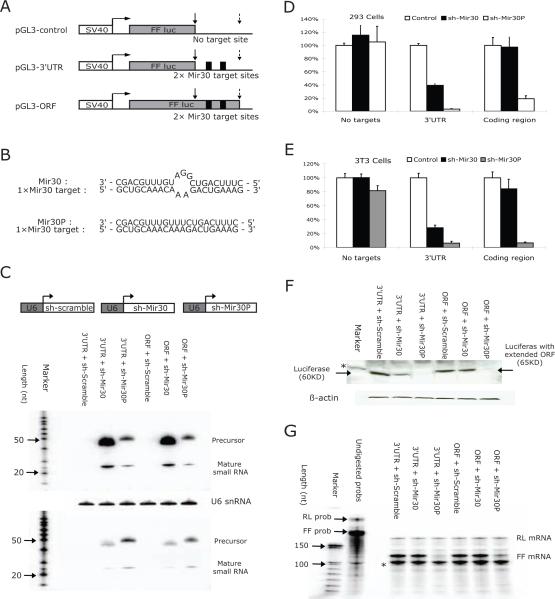
Figure 1. MiRNA-mediated repression is abolished in extended ORFs.
The structure of the reporter constructs used in this study. pGL3-control containing no miRNA target sites; pGL3-3'UTR with two tandem mir-30 targets sites located in 3' untranslated region; and pGL3-ORF with upstream stop-codon abolished and mir-30 target sites covered by extended ORF. Grey box represents the ORF of FF-luciferase gene. Dark box represents the tandem mir-30 target sites with six base-pairs in between. Positions of upstream (original) stop-codon and downstream stop-codon are indicated by solid and dotted arrows, respectively.
(A) Schematic illustration of the interactions between mir-30 target sequence and guiding strand sequence of sh-mir-30 and sh-mir-30P, respectively. (C) NIH3T3 cells were co-transfected with plasmids, as described above. Sh-RNA expressed from U6-driven cassette was detected by Northern blot using either a probe against mir-30 (Up) or a probe against mir-30P (Down). Due to sequence similarity, cross-hybridization was observed. Endogenous U6 snRNA was also detected as an internal control.
(D)HEK293 cells and (E) NIH3T3 cells were co-transfected with different combinations of plasmids, and dual-luciferase assays were performed 36hr post-transfection. FF-luciferase activities were normalized with RL-luciferase, and the percentage of relative enzyme activity compared to the negative control (treated with sh-scramble) was plotted. Error bars represent the standard deviation from three independent experiments, each performed in triplicate.
(F) Protein analysis by Western blot was performed in transfected 3T3 cells. A protein band of β-actin was used as an internal control. Positions of the bands representing wild-type or mutant FF-luciferase were indicated by arrows. A non-specific band was indicated by an asterisk. (G) RNA levels of either FF-luciferase or RL-luciferase from transected 3T3 cells were detected by ribonuclease protection assay (RPA). Full-length probes and protected bands were indicated in the figure. A band labeled with an asterisk is possibly due to a truncated RL probe and, therefore, corresponding to RL-luciferase mRNA level.