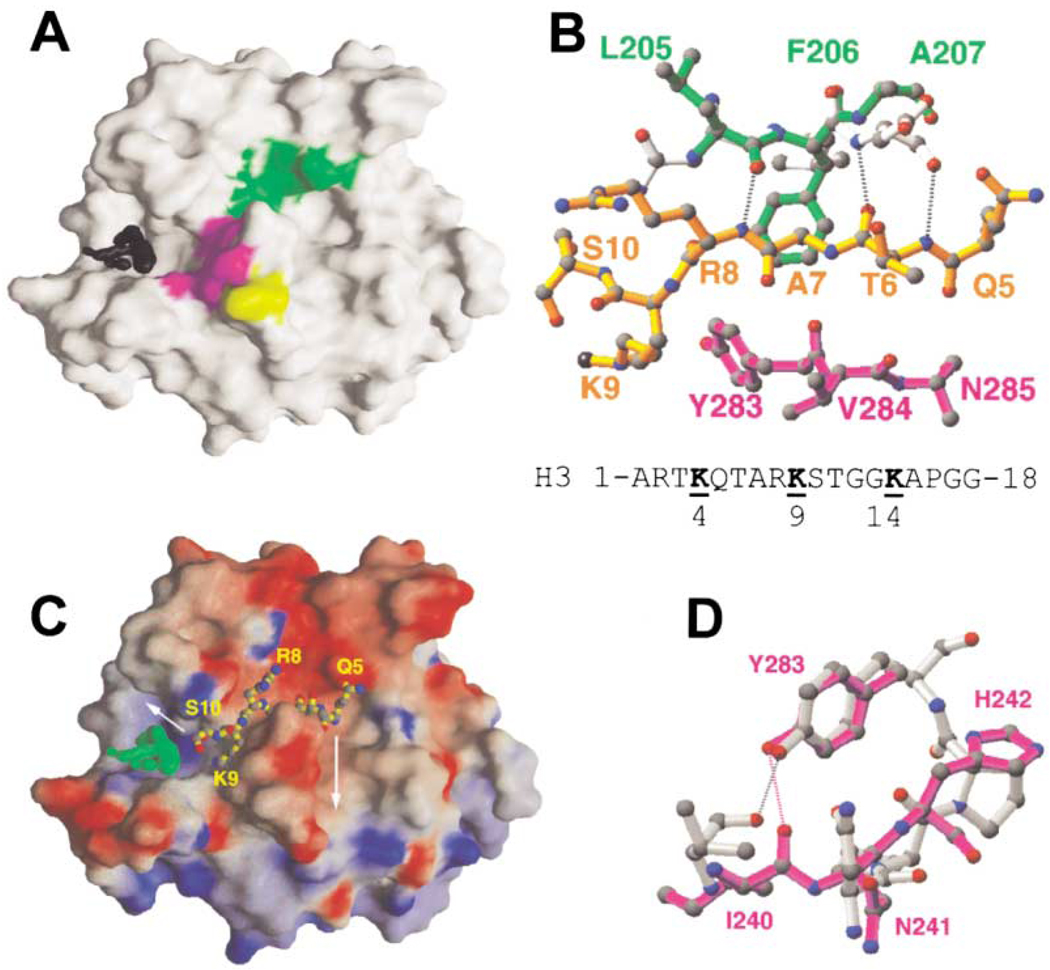
Figure 5. Putative Peptide Binding Cleft.
(A) Front view of GRASP surface (Nicholls et al., 1991). The difference electron density map (black) is contoured at 5.5σ. Strand β10 is green, N241, H242, and Y283 are magenta, and C244 is yellow.
(B) Superimposition of Drosophila HP1 β strand (light gray) (Jacobs and Khorasanizadeh, 2002; PDBcode 1KNA) and DIM-5 strand β10 (green). Dashed lines indicate the hydrogen bonds between HP1 and H3 peptide (yellow). The DIM-5 residues on the other side of the HP1 peptide are colored magenta. The dimethylated (methyl groups in black) target nitrogen atom occupies water site 2 (see Figure 4). The sequence of histone H3 peptide is shown at the bottom; both K4 and K14 are five residues away from K9.
(C) The docked H3 peptide lies in the putative peptide binding cleft. The cleft extends in both directions following turns as indicated. Surface charge distribution is displayed as blue for positive, red for negative, and white for neutral.
(D) Superimposition of active site NPPY residues of TaqI DNA-adenine amino MTase (gray) (Goedecke et al., 2001; PDB code 1G38) and the proposed DIM-5 active site residues N241, H242, and Y283 (magenta). The Tyr in both cases is hydrogen bonded to a main chain amide nitrogen atom (dashed bonds).