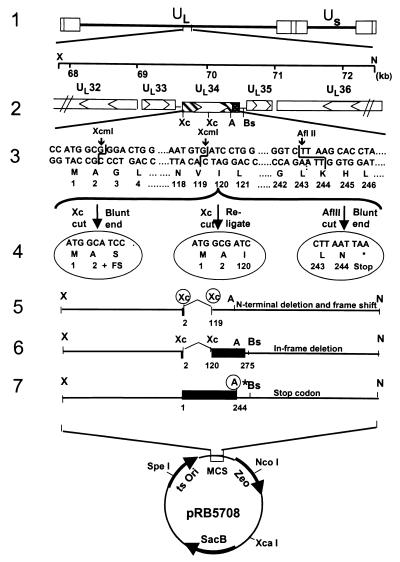
Figure 1.
Schematic representation of viral and plasmid DNA sequence arrangements. Line 1, A linear representation of the HSV-1 genome. The rectangles represent the inverted repeats flanking the unique sequences (UL and US represented by thin lines). Line 2, pRB5709, a 4.5-kb XhoI/NotI fragment of HSV-1(F) containing the UL33, UL34, UL35, and portion of UL32 and UL36 genes. The arrowheads indicate the direction of the transcription; solid dotted box indicates the putative transmembrane domain of the UL34 gene product. Line 3, DNA sequences of UL31 coding domains showing relevant restriction endonucleases sites used for construction of deletion mutants. Line 4, From left to right, pRB5710, pRB5711, and pRB5712. pRB5710 was derived by excision of a XcmI fragment in UL34, blunt-ending, and religation to produce a frame shift after the first two codons of UL34. pRB5711 was made by direct religation of the plasmid pRB5709 after removal of a XcmI fragment. This caused an in-frame deletion of codons 3–119. In pRB5712 the pRB5709 plasmid was cleaved with AflII, blunt-ended, and religated. In addition, a stop codon, and a unique restriction site (PacI), was introduced immediately after codon 244 of UL34. Lines 5–7, The transfer plasmids pRB5713, pRB5714, and pRB5715 were constructed by cloning the XhoI/NotI fragment from pRB5710, pRB5711, and pRB5712, respectively, into the transfer vector (pRB5708) at XhoI/NotI sites.