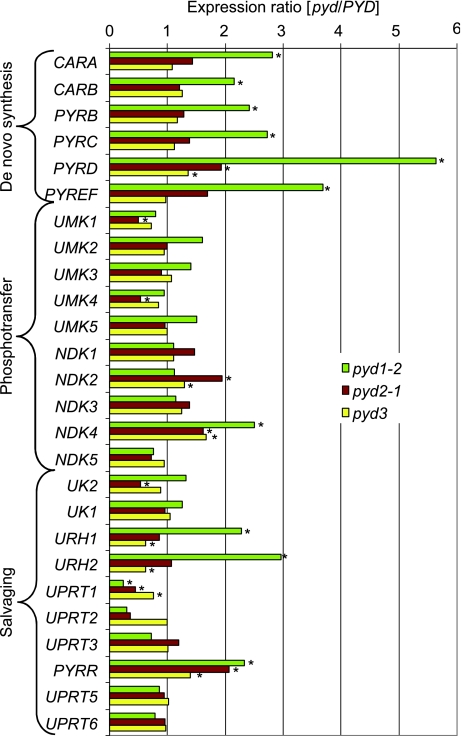
Fig. 6.
Relative transcript levels of pyrimidine nucleotide metabolism genes in pyd mutants. Arabidopsis seedlings (16-d-old) grown on plates containing half-strength Murashige and Skoog (MS) medium with 0.5% sucrose were analysed by quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR). Values are calculated as the difference of the Ct value from EF1α taken to the power of efficiency. Each bar represents the expression of the gene in question in the respective mutant relative to the expression in the wild-type. Each bar represents the mean values of six individual seedlings. Measurements were repeated twice. Significant differences (P < 0.05) using unpaired two-tailed t-tests are marked with an asterisk. CARA, carbamoylphosphate synthase, small subunit EC 6.3.5.5; CARB, carbamoylphosphate synthase, large subunit EC 6.3.5.5; NDK, nucleoside diphosphate kinase EC 2.7.4.6; PYRB, aspartate transcarbamoylase EC 2.1.3.2; PYRC, dihydroorotase EC 3.5.2.3; PYRD, dihydroorotate dehydrogenase EC 1.3.99.11; PYRFF, UMP synthase EC 2.4.2.10 and EC 4.1.1.23; PYRR, uracil phosphoribosyl transferase EC 2.4.2.9; UK, uridine kinase EC 2.7.1.48; UMK, uridine monophosphate kinase EC 2.7.4.4; UPRT, uracil phosphoribosyl transferase-like EC 2.4.2.9; URH, uridine nucleosidase EC 3.2.2.3.