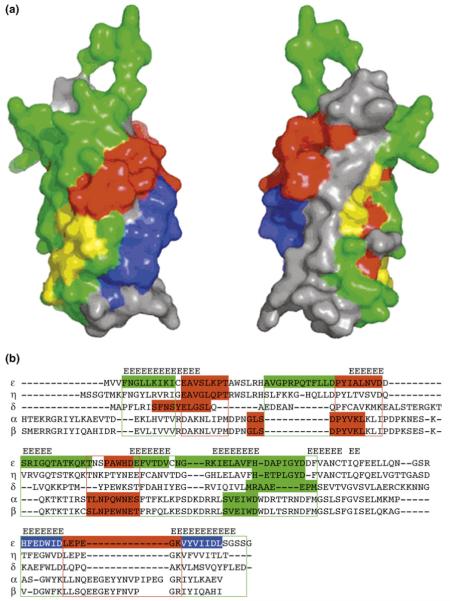
Figure 4.
Interaction cultures in the C2 domain of εPKC. (a) Space-filled surface mapping of the C2 domain of εPKC. The peptides derived from this domain and their activities are mapped back to the C2 domain of εPKC; they are colored in blue to represent PKC activators (not selective for εPKC), green to represent εPKC-selective activators, yellow to represent inconclusive results about their selectivity or red to represent εPKC inhibitors. εPKC-selective activators are peptides that cause phosphorylation of MARCKS in WT cells but not in εKO-cells and protect cardiac tissue from ischemia and reperfusion damage. Three of the four PKC-inhibiting peptides cluster to one area on the surface of the C2 domain (red regions). However, the selectivity of these inhibitory peptides for εPKC has not yet been determined. (b) Multiple sequence alignment of the C2 domain of five PKC isozymes identifies potential new peptides with similar biological activities derived from other isozymes. The alignment is based on structure (β strands are aligned) and sequence. Sequences corresponding to peptides with different biological activities are colored (red to represent inhibitors, green to represent selective activators or blue to represent isozyme non-selective activators). Peptides (indicated in color) from other isozymes are as follows: βPKC agonist ΨβRACK (SVEIWD) activates all the classical PKC isozymes [25], and βPKC antagonists βC2-4 (SLNPEWNET) and βC2-2 (MDPNGLSDPYVKL) are inhibitors of all the classical PKC isozymes [25]; δPKC agonist (δΨRACK, MRAAEDPM) and antagonist (δV1-1, SFNSYELGSL) are selective for δPKC [70], ηPKC agonist (ηΨRACK, HETPLGYD) and antagonist (ηV-2, EAVGLQPT) [67]. Sequence alignment of C2 domains of several PKC isozymes predicts regions from which potential peptide regulators (selective and nonselective) for other PKC isozymes can be generated (color, boxed areas). Reproduced, with permission, from Ref. [68].