Abstract
We have developed a rapid, straightforward, one plasmid dual positive/negative selection system for the evolution of aminoacyl-tRNA synthetases with altered specificities in E. coli. This system utilizes an amber stop codon containing a chloramphenicol acetyltransferase/uracil phosphoribosyltransferase fusion gene. We demonstrate the utility of the system by identifying a variant of the M. jannaschii tyrosyl synthetase from a library of 109 variants that selectively incorporates para-iodophenylalanine in response to an amber stop codon.
To date, more than 40 unnatural amino acids have been site-specifically incorporated into proteins directly in living organisms using orthogonal tRNA/aminoacyl-tRNA synthetase (aaRS) pairs.1–8 The majority of these have been incorporated in E. coli by directed evolution of an M. jannaschii tyrosyl-tRNA synthetase (MjYRS)/tRNACUA-derived pair which is orthogonal to the tRNAs and aaRSs in bacteria. The evolution process involves subjecting a library of plasmid-borne aminoacyl-tRNA synthetase variants, created by site-saturation mutagenesis of the amino acid binding pocket, to a series of positive and negative selections. The positive selection is carried out using a selection vector containing the amber suppressor tRNA and a chloramphenicol acteyltransferase (CAT) gene with an amber stop codon at a permissive site. Selection on chloramphenicol (Cm) in the presence of the unnatural amino acid enriches aaRS variants capable of incorporating either the unnatural amino acid or an endogenous amino acid. Negative selection utilizes a vector containing the tRNA and an arabinose-inducible gene encoding the toxic ribonuclease barnase with three amber stop codons at permissive sites; growth in the presence of arabinose eliminates those aaRS variants which recognize endogenous amino acids.1 A typical selection requires three rounds of positive and negative selection to obtain synthetase variants with the desired activity.
While the current selection system is effective, it has a number of disadvantages. First, the surviving pool of aaRS containing plasmids must be isolated after each selection step, and used to transform the appropriate host cells (harboring either the positive or negative selection vector) for the next step. Second, the stringency of the negative selection, while tunable to some extent by varying the concentration of arabinose, cannot be tuned over a broad dynamic range. Finally, since the selection system is encoded on two plasmids, changes to it for evolution of other tRNA/aaRS pairs or other components of the translational machinery, requires significant effort.
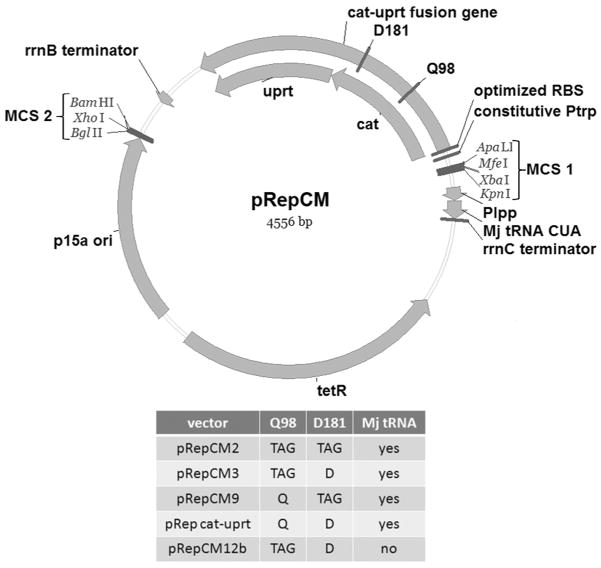
We have developed a new series of selection vectors (Figure 1) which overcome these limitations, allowing the rapid evolution of aminoacyl-tRNA synthetase specificity in bacteria. These vectors utilize a cat-uracil phosphoribosyltransferase (uprt) fusion gene, which has previously been shown to function as a dual positive/negative selection marker. UPRT converts 5-fluorouracil (5-FU) to 5-fluoro-dUMP, which inhibits thymidylate synthase, causing cell death.9 By inserting an amber stop codon at a permissive site in the cat portion of the fusion gene, amber suppression activity can be selected for in the presence of both Cm and an unnatural amino acid, and then selected against in the presence of 5-FU. We demonstrate that our dual selection vector can be used to identify an aminoacyl-tRNA synthetase mutant that selectively incorporates para-iodophenylalanine (pIPhe) from a library of 109 variants by a series of positive and negative selection steps without DNA isolation and retransformation. Cells are simply plated under selective conditions, harvested, and replated in the next step. Both positive and negative selections are tunable over a broad dynamic range by varying the concentrations of Cm and 5-FU, respectively. These selection vectors also contain two multi-cloning sites (MCS), allowing facile introduction of the tRNA and aaRS genes.
Figure 1.
Dual positive/negative selection vectors.
Four selection vectors (pRepCM2, 3, 9, 12b; Figure 1) containing either one or two amber codons at two different permissive sites (position 98, 181) of the cat gene, and a control vector (pRep cat-uprt) with no amber codons were constructed in several parallel steps. pRepCM12b lacks the MjtRNACUA, and was designed for the evolution of alternative tRNAs. The utility of pRepCM2, 3, and 9 for directed evolution of novel aaRS specificities was first tested by co-expression of each vector with wild-type M. jannaschii tyrosyl-tRNA synthetase, empty vector, and a previously evolved para-iodophenylalanine specific aaRS (pIPheRS)10 in the E. coli GH371 host strain, which has the genomic copy of the UPRT gene disrupted.9 To mimic positive selection conditions, each strain was assayed for growth on a range of Cm concentrations, and those strains harboring pIPheRS were assayed both in the presence and the absence of pIPhe (Figure 2A). All three selection vectors conferred aaRS-dependent, unnatural amino acid-dependent Cm resistance. As expected, the double amber stop codon construct, pRepCM2, had a lower maximum Cm resistance level than either of the single amber constructs. Interestingly, the D181TAG mutant (pRepCM9) showed a higher background amber suppression level than the Q98TAG mutant (pRepCM3). It is clear from these results that each construct is effective at selecting for mutants with the desired unnatural amino acid specificity, and against undesired mutants within a range of Cm concentrations. The optimal Cm concentration ranges for selection using the three constructs are as follows: 5–25 μg/mL for pRepCM2; 75–125 μg/mL for pRepCM3; pRepCM9 is not ideal for selection because its high background suppression narrows the effective dynamic range.
Figure 2.
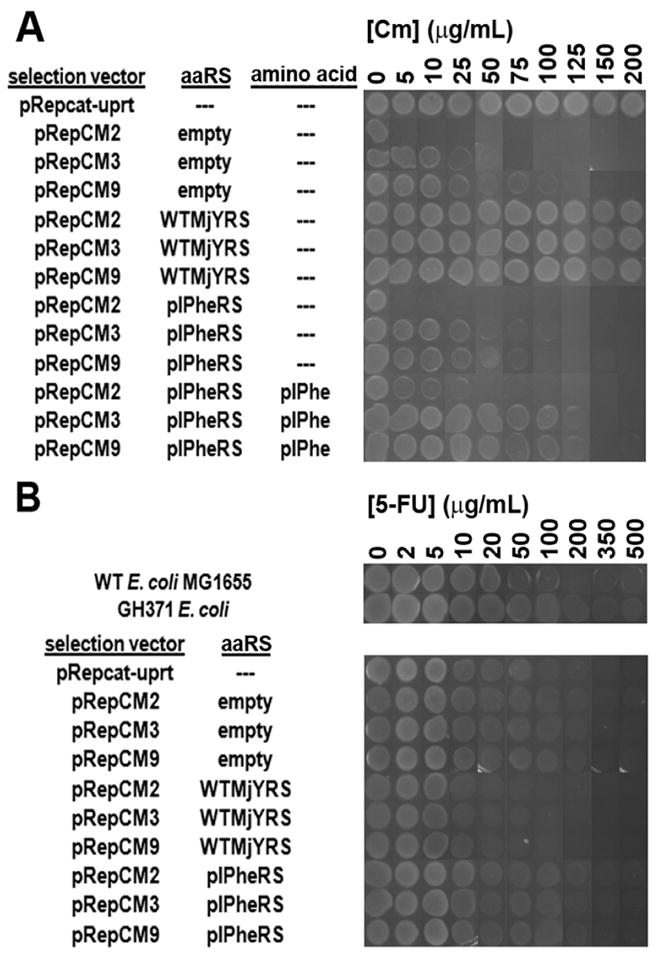
(A) Chloramphenicol resistance and (B) 5-fluorouracil sensitivity of dual positive/negative selection constructs pRepCM2 (Q98TAG, D181TAG), pRepCM3 (Q98TAG), and pRepCM9 (D181TAG) co-expressed with empty vector, wild-type MjYRS, or pIPheRS.
To test the constructs under conditions simulating a negative selection, each strain was assayed for growth on a range of 5-FU concentrations in the absence of pIPhe (Figure 2B). Each construct confers enhanced 5-FU sensitivity when co-expressed with wild-type MjTyrRS compared to co-expression with pIPheRS, indicating that synthetases incorporating endogenous amino acids can be selected against. The optimal 5-FU concentration range for selection is 100–200 μg/mL for all three constructs. The selection vector pRepCM3 was used for subsequent work because of its low background suppression and broad dynamic range.
Next, the one plasmid selection system was used to select an aminoacyl-tRNA synthetase that specifically incorporates pIPhe from a library of 109 naïve clones.11 The progress of the selection was monitored by sequencing ten random clones from the naïve library and from survivors after each selection step until complete convergence was observed. A different library12 than that originally used to identify the pIPheRS was used for this selection. As shown in Figure 3, the sequences converged on the published sequence for the pIPheRS after only three sequential selection steps. Furthermore, the effectiveness of the 5-FU negative selection can be seen by the increase in sequence convergence after the negative selection round. These results demonstrate that this one plasmid selection system is effective at identifying a specific unnatural amino acid incorporating synthetase variant from a large naïve library. Interestingly, positions 107 and 159, which were not randomized in this library, converged on non-wild type sequences identical to those seen in the original pIPheRS hit (E107S, I159L). Apparently these mutations were acquired at a low level either during library construction or by mutation during the selection.
Figure 3.
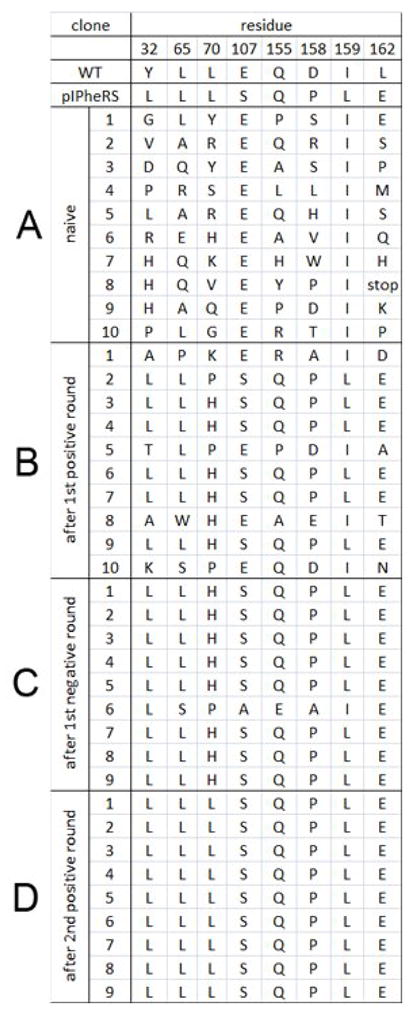
Sequences of clones selected randomly from (A) the naïve library, and from survivors after (B) the first positive round, (C) first negative round, and (D) second positive round.
In summary, we have developed a series of dual positive/negative selection vectors based on an amber stop codon-containing cat-uprt marker; optimized conditions for their use; and demonstrated their effectiveness for the evolution of aminoacyl-tRNA synthetases with novel specificities. These vectors were designed such that they can be easily adapted to a variety of selection schemes in which the readout is nonsense or frameshift suppression. The one plasmid selection system significantly reduces the amount of work required to carry out aaRS selections in E. coli, and will be a useful tool in future unnatural amino acid incorporation studies. This system complements a previously developed dual positive/negative selection system for aaRS selection in S. cerevisiae which relies on positive (H1S3) and negative (URA3) selectable markers under control of an amber nonsense mutation in the GAL4 transcription activator.13
Acknowledgments
We thank Jason W. Chin for providing plasmid p21-C9, which contains the cat-uprt fusion gene. This work is supported by NIH grants R01 GM062159 (P.G.S.) and F32 GM080067 (C.E.M.), and the Skaggs Institute for Chemical Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Xie J, Schultz PG. Nat Rev Mol Cell Biol. 2006;7:775. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 2.Liu C, Schultz PG. Nat Biotechnol. 2006;24:1436. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 3.Tippmann EM, Schultz PG. Tetrahedron. 2007;63:6182. [Google Scholar]
- 4.Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat Chem Biol. 2007;3:769. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 5.Guo J, Wang J, Anderson JC, Schultz PG. Ang Chem. 2008;47:722. doi: 10.1002/anie.200704074. [DOI] [PubMed] [Google Scholar]
- 6.Cellitti SE, Jones DH, Lagpacan L, Hao X, Zhang Q, Hu H, Brittain SH, Brinker A, Caldwell J, Bursulaya B, Spraggon G, Brock A, Ryu Y, Uno T, Schultz PG, Geierstanger BH. J Am Chem Soc. 2008;130:9268. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. Ang Chem. 2008;47:8220. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brustad E, Bushey ML, Brock A, Chittuluru J, Schultz PG. Bioorg Med Chem Lett. 2008;18:6004. doi: 10.1016/j.bmcl.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 9.Rackham O, Chin JW. Nat Chem Biol. 2005;1:159. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 10.Xie J, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. Nat Biotech. 2004;22:1297. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 11.The following selection conditions were used: GMML media, Cm 70 μg/mL, growth at 37 °C for 60–72 h for positive selection; LB media, 5-FU 125 μg/mL, growth at 37 °C for 20–24 h for negative selection. After each selection step the cells were scraped from the plate, quantified by OD600 measurement, and replated at an appropriate density.
- 12.Xie J, Liu W, Schultz PG. Ang Chem. 2007;46:9239. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 13.Chin JW, Cropp TA, Mukherji M, Zhang Z, Schultz PG. Science. 2003;10:511. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]