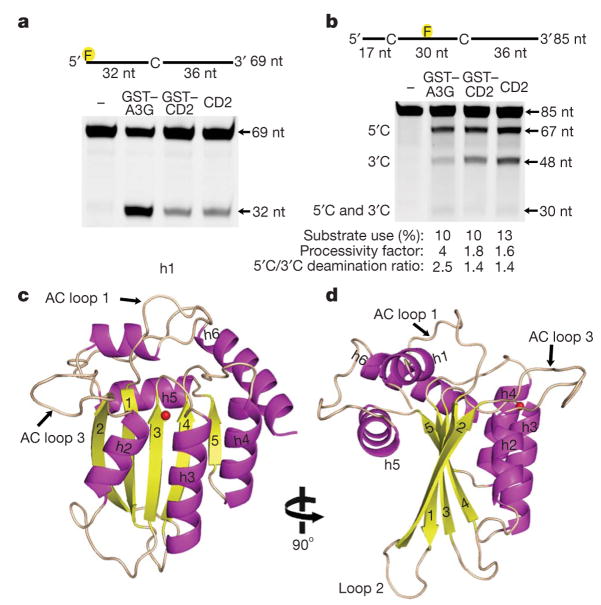
Figure 1. The X-ray structure of enzymatically active APOBEC3G-CD2.
a, Analysis of the deamination activity for full-length GST–APOBEC3G (GST–A3G) and GST–APOBEC3G-CD2 (GST–CD2) and APOBEC3G-CD2 (CD2). The 32-nucleotide (nt) band indicates deamination activity. b, APOBEC3G processivity and the 3′to 5′deamination bias was characterized on ssDNA with two CCC motifs. Single deaminations of the 5′C and the 3′C appear as 67- and 48-nucleotide fragments, respectively; deamination of both the 5′C and the 3′C results in a 30-nucleotide fragment (see Methods) c, d, Two views of the APOBEC3G-CD2 domain rotated 90° showing the five-stranded β-sheet core surrounded by six helices. The zinc is represented as a red sphere.