Abstract
Etoposide is routinely used in combination based chemotherapy for testicular cancer and small-cell lung cancer; however, myelosuppression, therapy-related leukemia and neurotoxicity limit its utility. To determine the genetic contribution to cellular sensitivity to etoposide, we evaluated cell growth inhibition in Centre d’ Etude du Polymorphisme Humain lymphoblastoid cell lines from 24 multi-generational pedigrees (321 samples) following treatment with 0.02–2.5 µM etoposide for 72 h. Heritability analysis showed that genetic variation contributes significantly to the cytotoxic phenotypes (h2 = 0.17–0.25, P = 4.9 × 10−5−7.3 × 10−3). Whole genome linkage scans uncovered 8 regions with peak LOD scores ranging from 1.57 to 2.55, with the most significant signals being found on chromosome 5 (LOD = 2.55) and chromosome 6 (LOD = 2.52). Linkage-directed association was performed on a subset of HapMap samples within the pedigrees to find 22 SNPs significantly associated with etoposide cytotoxicity at one or more treatment concentrations. UVRAG, a DNA repair gene, SEMA5A, SLC7A6 and PRMT7 are implicated from these unbiased studies. Our findings suggest that susceptibility to etoposide-induced cytotoxicity is heritable and using an integrated genomics approach we identified both genomic regions and SNPs associated with the cytotoxic phenotypes.
Introduction
Etoposide is one of the most widely used chemotherapeutic agents for the treatment of germ cell tumors, small-cell lung cancer, leukemia and lymphomas (Baldwin and Osheroff 2005; Kopp et al. 2006; Rigas and Lara 2005). Although its efficacy has been demonstrated as a single agent and in combination regimens, much of its anti-cancer benefits are limited by toxicities including myelosuppression, neurotoxicity and therapy related leukemia (Baldwin and Osheroff 2005).
The mechanism by which etoposide exerts its cytotoxic effect is via targeting topoisomerase II, specifically by both stabilizing and increasing the concentration of the topoisomerase II–DNA cleavage complex (Ross et al. 1984; Yang et al. 1985). While normally a short-lived intermediate within the catalytic cycle, an increased concentration of this complex within the cell leads to mutagenic effects or cell death, primarily by means of the apoptotic pathways (Lowe et al. 1993). Although it is well known that etoposide acts by interacting with topoisomerase II, the cellular processes and genetic variation associated with sensitivity to etoposide still remain largely unknown.
In previous studies, our lab (Dolan et al. 2004) and others (Watters et al. 2004) have used large CEPH pedigrees to demonstrate that drug-induced cytotoxicity is heritable and associated with one or more genomic loci using linkage analysis. Within the large CEPH pedigrees are International European (CEU) HapMap trios that have publicly available enriched genotypic information. Our lab utilized a linkage-directed association analysis to narrow down the chromosomal regions to a specific list of SNPs associated with daunorubicin induced cytotoxicity (Duan et al. 2007). In this report, lymphoblastoid cell lines (LCLs) derived from large CEPH pedigrees were used to identify the extent to which heritable factors contribute to etoposide-induced cytotoxicity. Whole genome linkage analysis was then followed by a linkage-directed association analysis to determine the extent to which SNPs within our linkage region contributed to the cytotoxic outcome.
Results
Cell cytotoxicity
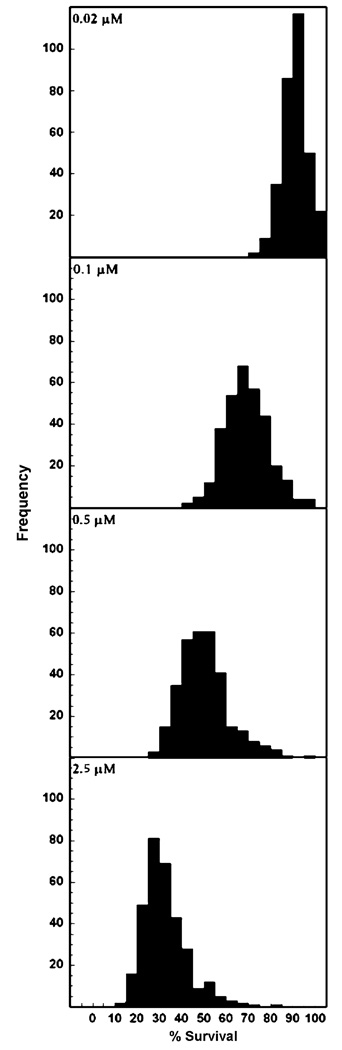
Using a short-term cell growth inhibition assay, we determined the cytotoxic effect of etoposide on 321 CEPH LCLs derived from 24 three-generation CEPH/Utah pedigrees. The median of the etoposide concentration required to inhibit 50% cellular growth (IC50) for all 321 cell lines following treatment with etoposide for 72 h was 0.4 µM. This value was in the lower range of those determined for a panel of NCI-60 human tumor cell lines (http://dtp.nci.nih.gov/) treated with etoposide for a longer time period, 96 h (range 0.2–63 µM, median 6.6 µM). As shown in Fig. 1, large variation in % survival for each concentration was observed. None of the five phenotypes (% cell survival at 0.02–2.5 µM etoposide and IC50) were normally distributed (P ≤ 0.05) based on Kolmogorov–Smirnov statistic. Therefore, phenotype data were transformed using the inverse normalization of the percentile rank function in Microsoft Excel™ software for further analysis.
Fig. 1.
Frequency of distribution plots of % survival for 321 CEPH cell lines after 72 h treatment with 0.02, 0.1, 0.5 and 2.5 µM etoposide
Heritability analysis
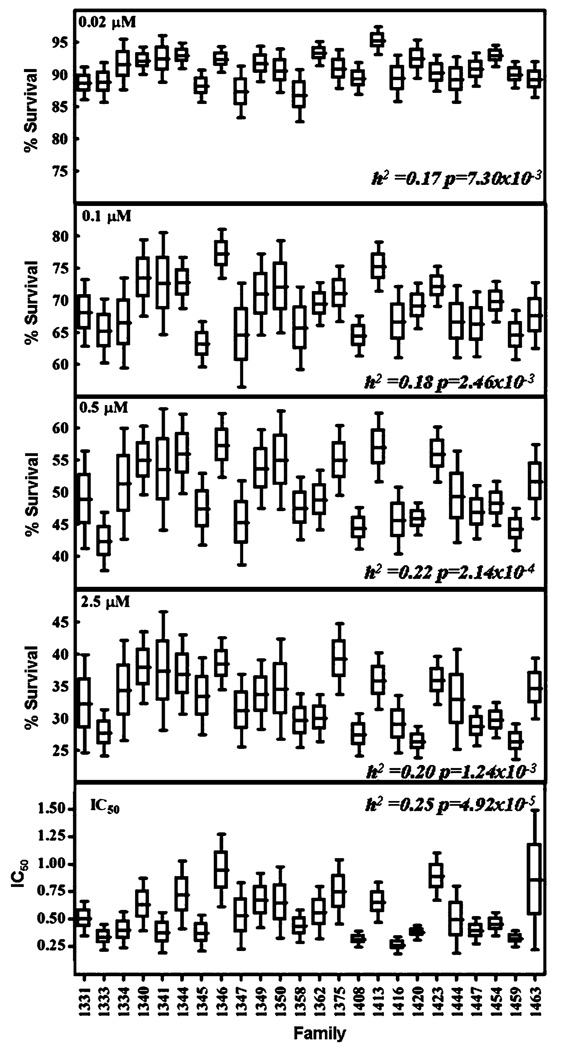
To quantify the proportion of genetic factors contributing to human LCL variation in sensitivity to etoposide, heritability analysis was performed. The variation of the cytotoxic phenotypes among the 24 CEPH families is illustrated in Fig. 2, with greater variation among families than within families. Sequential Oligogenic Linkage Analysis Routines (SOLAR) was used to calculate the heritability. As shown in Fig. 2, the heritability values for each concentration and IC50 range from 0.17 to 0.25, suggesting a significant genetic component contributing to the cytotoxic effect of etoposide. Age, sex and sex × age were tested as covariates, but none were significant.
Fig. 2.
Box plots for 24 pedigrees are shown for various concentrations and IC50 of etoposide illustrating inter and intra-family variance. The mean % survival and IC50 for each family following 72 h treatment with etoposide is represented by the line. Box represents mean ± SE, whiskers represent (mean ± 1.97 × SE). Heritability and level of significance for each phenotype is found in the plot
Linkage analysis
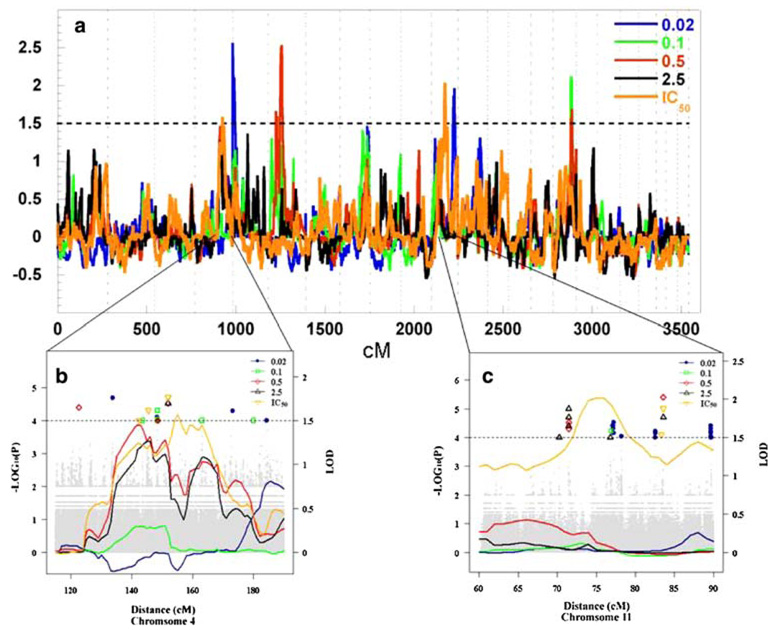
Non-parametric linkage analysis was performed on five phenotypes (0.02, 0.1, 0.5, 2.5 µM and IC50) using 7,209 heterozygous SNPs and microsatellite markers. Drug cytotoxicity is likely a multigenic trait, therefore LOD ≥ 1.5 was chosen in attempts to be inclusive of genes which have a moderate effect on drug-induced cytotoxicity. Using MERLIN multipoint analyses, eight genomic regions displayed suggestive LOD scores of ≥ 1.5 on five chromosomes (Fig. 3a; Table 1). The highest LOD score (2.55) was seen following 0.02 µM etoposide treatment and located on chromosome 5. Peaks on two chromosomal regions (chromosome 5 and 16) showed an inverse relationship between LOD score and concentration of drug tested indicative of genetic variants contributing to a greater extent at lower concentrations of etoposide in these regions (data not shown).
Fig. 3.
Genome wide linkage analysis scan and concentration dependent peaks of interest. a Genome wide linkage scans of each phenotypic response for 321 LCLs representing 24 CEPH families. Dashed horizontal line denotes LOD = 1.5; etoposide concentrations: 0.02 µM (blue), 0.1 µM (green), 0.5 µM (red), 2.5 µM (black), IC50 (orange). b and c Focused view of linkage peaks with significant results of QTDT analysis using genotypes for 86 CEU cell lines from the HapMap Project and associated genotypes on (b) chromosome 4 (c) chromosome 11. Horizontal dashed line P = 1 × 10−4; LOD = 1.5) (0.02 µM, blue dots; 0.1 µM, green dots; 0.5 µM, red dots; 2.5 µM, black dots; IC50, orange dots). The gray dots are non-significant results of QTDT analysis
Table 1.
Location of linkage regions with LOD ≥ 1.5
| Phenotype | Chromosome | LOD score |
1-LOD range (cM) |
Number of SNPs within 1-LOD range |
|---|---|---|---|---|
| IC50 | 4 | 1.57 | 132–175 | 26,802 |
| 0.02 µM | 5 | 2.55 | 0–14 | 13,520 |
| 0.5 µM | 6 | 1.65 | 43–63 | 13,769 |
| 0.5 µM | 6 | 2.52 | 67–83 | 8,140 |
| IC50 | 11 | 2.02 | 59–91 | 19,570 |
| 0.02 µM | 11 | 1.95 | 118–136 | 13,870 |
| 0.1 µM | 16 | 2.11 | 99–112 | 4,042 |
| 0.5 µM | 16 | 1.68 | 99–117 | 7,497 |
Association analysis
Association tests were performed between 103,168 common SNPs (minor allele frequency >5%) that were within the one LOD confidence intervals for the significant linkage peaks (LOD ≥ 1.5) of all five chromosomes and the corresponding cytotoxic phenotype(s) at which a significant linkage signal was detected. Another words, association studies were performed using the same phenotype (% survival following treatment with a given etoposide concentration) as the phenotype of the significant linkage region. Figure 3b, c display SNPs found significant within our identified linkage region on chromosome 4 and 11. We identified 22 unique SNPs significantly associated with cytotoxic phenotypes, of which 13 SNPs associated with percentage survival following 0.02 µM etoposide treatment, 3 SNPs with that of 0.5 µM treatment and 6 SNPs with the IC50 phenotype (Table 2). Sixteen of 22 SNPs were located within 4 known gene regions, the remaining 6 SNPs resided within intergenic regions of the genome (P ≤ 1 × 10−4, false discovery rate (FDR) = 0.17 ~ 0.60). Four genes with significant associated SNPs were found distributed among chromosomes 5, 11 and 16. No significant associations were found between genetic markers in the linkage peak of chromosome 6 associated with survival at 0.5 µM etoposide.
Table 2.
Linkage directed association results
| SNP | Genomic region | cM | Phenotype | Local loci | Functional class |
|---|---|---|---|---|---|
| rs17232998 | chr4:128540333 | 142.3445 | IC50 | – | |
| rs13110934 | chr4:131343379 | 145.4486 | IC50 | – | |
| rs9990901 | chr4:137147902 | 151.8765 | IC50 | – | |
| rs17048403 | chr4:137171324 | 151.9024 | IC50 | – | |
| rs1863984 | chr5:4264882 | 4.614959 | 0.02 µM | – | |
| rs10079862 | chr5:9512730 | 10.29357 | 0.02 µM | SEMA5A | Intron |
| rs571826 | chr5:9513453 | 10.29435 | 0.02 µM | SEMA5A | Intron |
| rs16882871 | chr5:9532622 | 10.31509 | 0.02 µM | SEMA5A | Intron |
| rs10213926 | chr5:9535547 | 10.31826 | 0.02 µM | SEMA5A | Intron |
| rs2135071 | chr5:9536171 | 10.31893 | 0.02 µM | SEMA5A | Intron |
| rs3777359 | chr5:9543419 | 10.32678 | 0.02 µM | SEMA5A | Intron |
| rs369459 | chr5:9560624 | 10.34539 | 0.02 µM | SEMA5A | Intron |
| rs446732 | chr5:9560772 | 10.34555 | 0.02 µM | SEMA5A | Intron |
| rs421548 | chr5:9561979 | 10.34686 | 0.02 µM | SEMA5A | Intron |
| rs442173 | chr5:9562018 | 10.3469 | 0.02 µM | SEMA5A | Intron |
| rs486947 | chr5:9562837 | 10.34779 | 0.02 µM | SEMA5A | Intron |
| rs268478 | chr5:9563136 | 10.34811 | 0.02 µM | SEMA5A | Intron |
| rs7116263 | chr11:75404436 | 83.29303 | IC50 | UVRAG | Intron |
| rs2510733 | chr11:75644259 | 83.55795 | IC50 | – | |
| rs11644360 | chr16:66884590 | 101.8842 | 0.5 µM | SLC7A6 | Intron |
| rs1127773 | chr16:66890248 | 101.8928 | 0.5 µM | SLC7A6 | 3′-UTR |
| rs3785125 | chr16:66945893 | 101.9775 | 0.5 µM | PRMT7 | Intron |
Discussion
Identification of genetic variants that predict drug response is an important component of personalized medicine (Hartford and Dolan 2007; Katoh and Katoh 2004; Ooyama et al. 2007). In the current study, our objective was to ascertain the component of etoposide-induced cytotoxicity due to genetics with the long-term goal of identifying genetic variation that identifies patients at risk for toxicity. Etoposide-induced cytotoxicity was determined to be a heritable trait with genetics contributing to 17–25% of the variation. Because the cell lines were derived from individuals within large pedigrees, whole genome linkage analysis was utilized to identify regions that harbored genetic variation important in sensitivity to etoposide. This list was further narrowed down by performing association studies with SNPs within the linkage regions (LOD ≥ 1.5) on a subset of densely genotyped HapMap samples (n = 86) and the cytotoxic phenotype(s) for which a QTL signal was detected. The final list of 22 SNPs identified through this linkage-directed association study are all considered important and may play a role in chemosensitivity through effects on gene expression, function or protein stability.
With enriched resources of markers and SNP genotypic information (http://www.cephb.fr/cephdb/ and http://www.hapmap.org) available, the LCL system has been used in mapping of genetic variants influencing other complex traits. Previously, cell response to ionizing radiation induced stress (Jen and Cheung 2003), cytotoxicity (Dolan et al. 2004; Watters et al. 2004) and variation in mRNA expression level (Morley et al. 2004). Studies performed on cisplatin (Dolan et al. 2004), 5-FU and docetaxel (Watters et al. 2004) and daunorubicin (Duan et al. 2007) have demonstrated that drug-induced cytotoxicity is heritable and amenable to genetic dissection. We compared the linkage peaks with those in our previous studies of daunorubicin (Duan et al. 2007) and cisplatin (Shukla et al. 2008). Interestingly, the results show there are two peaks that overlap with other drug phenotypes (LOD ≥ 1.5). The first located at chromosome 4 (132–175 cM) for etoposide (IC50) overlaps with daunorubicin (4q28.2–32.3 for survival at 0.025, 0.05, 0.1, 0.2 µM daunorubicin and IC50) and cisplatin (4q21.3–q35.2 for survival at 1, 5, 20 µM drug). Another peak is located at chromosome 16 (99–112 cM) for etoposide 0.1 and 0.5 µM that overlaps with both daunorubicin (16q23.1–24.1 for survival at 0.0125 µM drug) and cisplatin (16q23.1–q24.1 at 20 µM drug). However, our association results for those overlapping regions did not show common significant SNP associations between the three drugs. This discordance may be explained by a lack of power in the association studies due to the limited sample size of the 30 HapMap trios or possibly because the genotypes contributing to drug-induced cytotoxicity are not yet in HapMap. Although the same 24 large CEPH pedigrees were evaluated for daunorubicin, an additional 10 pedigrees were evaluated for cisplatin.
Etoposide is known as an inhibitor of the enzyme topoisomerase II that controls and alters the topologic states of DNA during transcription (Ross et al. 1984; Yang et al. 1985). In the present linkage and association studies, we failed to find significant association results for the topoisomerase II and other transporter genes that are involved in drug sensitivity. It is noteworthy that a candidate gene, GSTP1 is found in the chromosome 11 linkage peak (59–91 cM) with etoposide, although none of the SNPs in the GSTP1 locus shows significant association with etoposide (data not shown). Our linkage-directed association analysis resulted in the identification of 4 genes (SEMA5A, SLC7A6, PRMT7 and UVRAG) among three chromosomes (5, 11 and 16) harboring SNPs significantly associated with sensitivity to etoposide.
As shown in Table 2, a total of 12 high linkage disequilibrium SNPs in the intronic region of SEMA5A gene are shown to be associated with survival after treatment with 0.02 µM etoposide. SEMA5A is a member of the semaphorin protein family which is involved in axonal guidance during neural development (Adams et al. 1996). Previously, we also found two axon guidance pathway genes (NGEF and SLIT3) associated with daunorubicin induced cytotoxicity. We correlated the significant SNPs with the gene expression level as described previously (Huang et al. 2007a) and found that the SNPs (best SNP: rs369459, P = 3 × 10−7) in the SEMA5A gene loci were distantly associated with the expression of AMOT gene. AMOT is a member of angiostatin binding proteins that inhibit endogenous angiogenesis in vivo (Aase et al. 2007). In addition, two SLC7A6 SNPs and one PRMT7 SNP were found significantly associated with cell survival at 0.5 µM etoposide. SLC7A6 encodes an amino acid transporter. Expression level of SLC7A6 protein is involved in nitric oxide synthesis (Arancibia-Garavilla et al. 2003) that ultimately induces apoptosis and a series of other important biological processes (Leon et al. 2008). PRMT7 is an arginine methyltransferase catalyzing an irreversible protein modification (Miranda et al. 2004). PRMT family members were found to be involved in a variety of processes (Bedford and Richard 2005). Down-regulation of PRMT7 enzyme sensitizes tumor cells to camptothecin (Verbiest et al. 2008) and DNA damaging agents (Gros et al. 2006). Our data supports the role of PRMT7 in cellular sensitivity to etoposide.
A polymorphic site (rs7116263, C/G) was found within the intronic region of the gene UVRAG. This gene is implicated in resistance to damage induced by ultraviolet radiation as well as a positive regulator of Beclin-1 mediated autophagy (Liang et al. 2007, 1999). By forming a complex with the phosphatidylinositol 3-kinase C3 (PI3KC3), Beclin-1 initiates the early stage production of autophagosomes. Differential interaction of Beclin-1 with BCL-2 in both normal and autophagic conditions shows suppression of autophagic activity (Pattingre et al. 2005). Liang et al. (2006) performed detailed binding studies demonstrating that UVRAG and Beclin-1 directly interact and that Beclin-1 functions as a platform for the formation of the Bcl-2-UVRAG-PI3KC3 complex. A reduced level of Beclin-1 results in less UVRAG-induced autophagosome formation and vice versa (Liang et al. 2006). Interestingly, recent evidence has implicated autophagy through Beclin-1 in CaSki cervical carcinoma cells treated with etoposide (Lee et al. 2007). Our whole genome approach identifies a SNP within UVRAG associated with etoposide cytotoxicity. Further research on the role of this gene in sensitivity to etoposide is warranted.
Publicly available gene expression and genotypic data makes LCLs a potential resource to build models for identifying how genetic variation aVects susceptibility to drugs. Nonetheless, there are limitations with these cell lines. One limitation is the association studies in these cells is dependent on the SNPs identified through HapMap, thus they may not be the causal SNP but a marker in linkage disequilibrium. As more genetic markers are deposited, the linkage-directed association studies can be re-evaluated. In addition, the large CEPH pedigrees are derived from healthy Caucasian individuals of European descendants; hence results may not apply to individuals of different ethnicities or even populations that experience severe chemotherapeutic toxicity. LCLs may not represent a phenotypic effect that occurs in a specific tissue (e.g., neurotoxicity). Finally, although EBV transformation could introduce phenotypic or gene expression changes, the ability to map the expression phenotype (Cheung et al. 2005; Morley et al. 2004) provides some level of confidence in the model for genetic studies because gene expression is genetically controlled. Despite these limitations, LCLs provide a comprehensive, unbiased model to discover pharmacogenomic signatures of chemotherapeutic agents that are not possible in humans.
Using a linkage-directed association approach with drug induced cytotoxicity allowed for the identification of genes and SNPs contributing to human variation in sensitivity to etoposide. These findings may serve as a platform to further explore and improve our understanding of mechanisms of etoposide induced cytotoxicity and provides a list of potential genetic markers that may be further evaluated in clinical studies.
Materials and methods
Cell lines
Lymphoblastoid cell lines derived from 24 Caucasian Utah CEPH families (1331, 1333, 1334, 1340, 1341, 1344, 1345, 1346, 1347, 1349, 1350, 1358, 1362, 1375, 1408, 1413, 1416, 1420, 1423, 1444, 1447, 1454, 1459 and 1463) were purchased from the Coriell Institute for Medical Research (Camden, NJ). Cell lines were maintained in RPMI 1640 media (Mediatech, Herndon, VA) supplemented with 15% fetal bovine serum (HyClone, Logan, UT) and 1% l-glutamine (Invitrogen, Carlsbad, CA). Cell lines were diluted 3 times per week to a concentration 350,000 cells/ml and maintained at a 37°C, 5% CO2 and 95% humidity.
Drug
Etoposide (NSC-141540) was kindly provided by the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, National Cancer Institute, Bethesda, MD.
Cell cytotoxicity assay
Cell growth inhibition was evaluated following increasing concentrations of etoposide treatment (0, 0.02, 0.1, 0.5 and 2.5 µM for 72 h). Etoposide was prepared in 0.05% DMSO/phosphate buffered saline immediately before use as described previously (Huang et al. 2007b). Cell growth inhibition was determined using the non-toxic colorimetric based assay, alamarBlue® (Biosource, Camarillo, CA) as described (Duan et al. 2007). Briefly, cells with viabilities ≥85% were plated at a density of 1 × 105 cells/ml (1 × 104 cells per well), in triplicate, in 96-well round bottom plates. Following 24 h incubation, cells were treated with either vehicle (media contains 0.05% DMSO) or increasing concentrations of etoposide for 72 h.
Heritability and linkage analysis
To estimate the power to detect heritability, we used Genometric Analysis Simulation Program (GASP) (http://research.nhgri.nih.gov/gasp/) to simulate a quantitative phenotype with varying heritabilities in 24 CEPH families each with 15 members. For each replicate (100 replicates were simulated), we calculated the heritability with SOLAR. Power was estimated to be the proportion of replicates in which the heritability was significantly different from 0 (P < 0.05). We had 83% power to detect a heritability of at least 0.15 in this analysis. Heritability analysis was performed using SOLAR to estimate narrow sense heritability (h2) and test its significance at each treatment concentration as described previously (Duan et al. 2007). Briefly, all phenotype data were transformed using the inverse normalization of the percentile rank function in Microsoft Excel™ software. Age, sex, and age × sex interaction were used as covariates. Non-parametric linkage analysis was performed using MERLIN (Abecasis et al. 2002) as described previously (Duan et al. 2007). The genotypic data and map distances were downloaded from the CEPH Version 9 database and the Marshfield map database using error-checked markers.
Error checking
Error checking for Mendelian incompatibility, misspecified relationships and unlikely recombinations have been performed as described previously (Dolan et al. 2004), however, this study utilized a much denser map. The web-based platform integrates and formats data (pedigree, genotype, phenotype), executes error checking using PedCheck (O’Connell and Weeks 1998) to detect genotypic incompatibilities, PREST (Sun et al. 2002) to detect relationship misspecifications and multipoint engine for rapid likelihood inference (MERLIN) (Abecasis et al. 2002) to detect unlikely recombinants prior to linkage analysis and is enabled to run linkage analysis on multiple platforms including MERLIN, GENEHUNTER and SOLAR. From the combined pool of genotyped markers, 7,209 SNP and microsatellite non-redundant markers yielding a very dense genetic map with highly heterozygous markers (heterozygosity: 1% at <0.7, 7% at 0.7–0.8, 28% at 0.8–0.9, 64% at 0.9–1) were utilized for linkage mapping studies.
SNPs associated with etoposide-induced cytotoxicity
The 90 HapMap CEU cell lines were a component of the 24 large pedigrees we evaluated for cytotoxicity. Four samples (GM11839, GM12716, GM12717 and GM12236) were not included in the analysis due to the low viability or unavailability from Coriell at the time of phenotyping. As shown in Table 1, a total of 103,168 SNPs within the 1 LOD confidence intervals of linkage regions with LOD scores ≥1.5 were retrieved from the HapMap database (release 22). Only the SNPs without Mendelian transmission errors and minor allele frequency ≥5% were included in the follow-up association analysis. Association between SNPs and percentile rank transformed phenotypes (the cytotoxic phenotype(s) for which a QTL signal was detected) was performed using the QTDT program (Abecasis et al. 2000a, b). Gender was used as a covariate. P ≤ 0.0001 was considered statistically significant. FDR was used to control for multiple testing within each cytotoxic phenotype using the q value package in R statistics software (Storey and Tibshirani 2003).
Acknowledgments
We are extremely grateful to Dr. Jeong-Ah Kang for maintaining the cell lines. This Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org) study was supported by NIH/NIGMS grant GM61393 with data deposits supported by U01GM61374. Phenotypic data reported in this article can be accessed through http://www.PharmGKB.org by using data accession number PS207137.
Abbreviations
- EBV
Epstein–Barr Virus
- LCLs
Lymphoblastoid cell lines
- CEPH
Center d’Etude du Polymorphisme Humain
- CHR
Chromosome
- SNPs
Single nucleotide polymorphisms
- SOLAR
Sequential Oligogenic Linkage Analysis Routines
- MERLIN
Multipoint Engine for Rapid Likelihood Inference
- QTL
Quantitative trait loci
- FDR
False discovery rate
- QTDT
Quantitative transmission disequilibrium test
- LOD
Logarithm of the odds
Appendix: web resources
The URLs for data presented herein are as follows:
Pharmacogentics of Anticancer Agents Research Group (PAAR), http://www.pharmacogenetics.org
Pharmacogenetics Research Network and Database, http://www.pharmgkb.org
CEPH database, http://www.cephb.fr/cephdb/
International HapMap Project, http://www.hapmap.org
Coriell Institute for Medical Research, http://www.coriell.org
Marshfield map database, http://research.marshfieldclinic.org/genetics/home/index.asp
The SNP Consortium, http://www.snp.cshl.org/
NCBI dbGENE, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
R statistics software, http://www.r-project.org
Contributor Information
Wasim K. Bleibel, Department of Medicine, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL 60637, USA
Shiwei Duan, Department of Medicine, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL 60637, USA.
R. Stephanie Huang, Department of Medicine, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL 60637, USA.
Emily O. Kistner, Department of Health Studies, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL, USA
Sunita J. Shukla, Department of Human Genetics, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL, USA
Xiaolin Wu, Department of Medicine, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL 60637, USA.
Judith A. Badner, Department of Psychiatry, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL, USA
M. Eileen Dolan, Email: edolan@medicine.bsd.uchicago.edu, Department of Medicine, University of Chicago, 5841 S. Maryland Ave, Box MC2115, Chicago, IL 60637, USA.
References
- Aase K, Ernkvist M, Ebarasi L, Jakobsson L, Majumdar A, Yi C, Birot O, Ming Y, Kvanta A, Edholm D, Aspenstrom P, Kissil J, Claesson-Welsh L, Shimono A, Holmgren L. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–2068. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000a;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000b;8:545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Adams RH, Betz H, Puschel AW. A novel class of murine semaphorins with homology to thrombospondin is differentially expressed during early embryogenesis. Mech Dev. 1996;57:33–45. doi: 10.1016/0925-4773(96)00525-4. [DOI] [PubMed] [Google Scholar]
- Arancibia-Garavilla Y, Toledo F, Casanello P, Sobrevia L. Nitric oxide synthesis requires activity of the cationic and neutral amino acid transport system y + L in human umbilical vein endothelium. Exp Physiol. 2003;88:699–710. doi: 10.1113/eph8802647. [DOI] [PubMed] [Google Scholar]
- Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Badner JA, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros L, Renodon-Corniere A, de Saint Vincent BR, Feder M, Bujnicki JM, Jacquemin-Sablon A. Characterization of prmt7alpha and beta isozymes from Chinese hamster cells sensitive and resistant to topoisomerase II inhibitors. Biochim Biophys Acta. 2006;1760:1646–1656. doi: 10.1016/j.bbagen.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Hartford CM, Dolan ME. Identifying genetic variants that contribute to chemotherapy-induced cytotoxicity. Pharmacogenomics. 2007;8:1159–1168. doi: 10.2217/14622416.8.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, Schweitzer AC, Blume JE, Dolan ME. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007a;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol Cancer Ther. 2007b;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Pharmacogenomics on gastric cancer. Cancer Biol Ther. 2004;3:566–567. doi: 10.4161/cbt.3.6.936. [DOI] [PubMed] [Google Scholar]
- Kopp HG, Kuczyk M, Classen J, Stenzl A, Kanz L, Mayer F, Bamberg M, Hartmann JT. Advances in the treatment of testicular cancer. Drugs. 2006;66:641–659. doi: 10.2165/00003495-200666050-00005. [DOI] [PubMed] [Google Scholar]
- Lee SB, Tong SY, Kim JJ, Um SJ, Park JS. Caspase-independent autophagic cytotoxicity in etoposide-treated CaSki cervical carcinoma cells. DNA Cell Biol. 2007;26:713–720. doi: 10.1089/dna.2007.0577. [DOI] [PubMed] [Google Scholar]
- Leon L, Jeannin JF, Bettaieb A. Post-translational modifications induced by nitric oxide (NO): implication in cancer cells apoptosis. Nitric Oxide. 2008;19:77–83. doi: 10.1016/j.niox.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–22907. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooyama A, Okayama Y, Takechi T, Sugimoto Y, Oka T, Fukushima M. Genome-wide screening of loci associated with drug resistance to 5-fluorouracil-based drugs. Cancer Sci. 2007;98:577–583. doi: 10.1111/j.1349-7006.2007.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Rigas JR, Lara PN., Jr Current perspectives on treatment strategies for locally advanced, unresectable stage III non-small cell lung cancer. Lung Cancer. 2005;50(Suppl 2):S17–S24. [PubMed] [Google Scholar]
- Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984;44:5857–5860. [PubMed] [Google Scholar]
- Shukla SJ, Duan S, Badner JA, Wu X, Dolan ME. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- Verbiest V, Montaudon D, Tautu MT, Moukarzel J, Portail JP, Markovits J, Robert J, Ichas F, Pourquier P. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rowe TC, Liu LF. Identification of DNA topoisomerase II as an intracellular target of antitumor epipodophyllotoxins in simian virus 40-infected monkey cells. Cancer Res. 1985;45:5872–5876. [PubMed] [Google Scholar]