Table 1. Crystallographic data and refinement statistics.
Values in parentheses are for the last shell.
| PPARα–TIPP-703 | PPARγ–TIPP-703 | PPARδ–TIPP-401 | PPARδ–TIPP-204 | |
|---|---|---|---|---|
| Pan agonist | Pan agonist | α,δ dual | δ-specific | |
| Data collection | ||||
| Space group | P21 | C2 | P21 | P21 |
| Unit-cell parameters | ||||
| a (Å) | 44.372 | 93.307 | 39.492 | 39.172 |
| b (Å) | 61.529 | 61.604 | 93.149 | 91.947 |
| c (Å) | 53.124 | 118.973 | 96.370 | 96.361 |
| β (°) | 106.290 | 103.640 | 97.480 | 98.010 |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | 1.00000 |
| Resolution (Å) | 35.0–2.00 (2.07–2.00) | 50.0–2.40 (2.49–2.40) | 50.0–3.00 (3.11–3.00) | 50.0–2.65 (2.74–2.65) |
| No. of unique reflections | 18142 (1646) | 25166 (2065) | 13670 (1178) | 19488 (1827) |
| Completeness (%) | 98.7 (90.8) | 97.1 (80.3) | 97.8 (84.9) | 99.0 (93.3) |
| I/σ(I) | 10.6 (3.3) | 15.3 (2.7) | 7.7 (2.1) | 8.6 (2.7) |
| Redundancy | 3.7 (3.1) | 3.5 (2.8) | 3.7 (2.9) | 3.7 (3.1) |
| Rmerge† (%) | 6.4 (22.2) | 4.1 (26.1) | 8.8 (29.7) | 9.3 (28.2) |
| Refinement | ||||
| Resolution range (Å) | 35.0–2.00 | 50.0–2.40 | 38.0–3.00 | 42.4–2.65 |
| Rwork‡/Rfree§ | 21.4/25.3 | 24.0/28.6 | 23.0/28.8 | 21.4/27.6 |
| No. of atoms | ||||
| Protein | 2054 | 4111 | 4198 | 4215 |
| Water | 146 | 46 | 5 | 83 |
| Ligand | 37 | 66 | 113 | 98 |
| Average B factor (Å2) | ||||
| Protein | 24.74 | 49.32 | 42.46 | 31.73 |
| Water | 29.68 | 41.22 | 43.66 | 30.02 |
| Ligand | 28.45 | 65.85 | 25.22 | 31.76 |
| R.m.s.d. | ||||
| Bond lengths (Å) | 0.008 | 0.010 | 0.010 | 0.008 |
| Angles (°) | 1.2 | 1.3 | 1.4 | 1.3 |
| PDB code | 2znn | 2zno | 2znp | 2znq |
R
merge = 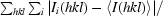
 , where 〈I(hkl)〉 is the mean I(hkl) over symmetry-equivalent reflections.
, where 〈I(hkl)〉 is the mean I(hkl) over symmetry-equivalent reflections.
R
work = 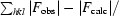
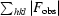 , where F
obs and F
calc are the observed and calculated structure factors, respectively.
, where F
obs and F
calc are the observed and calculated structure factors, respectively.
R free was calculated using 5% of the total reflections, which were chosen randomly and omitted from the refinement.
