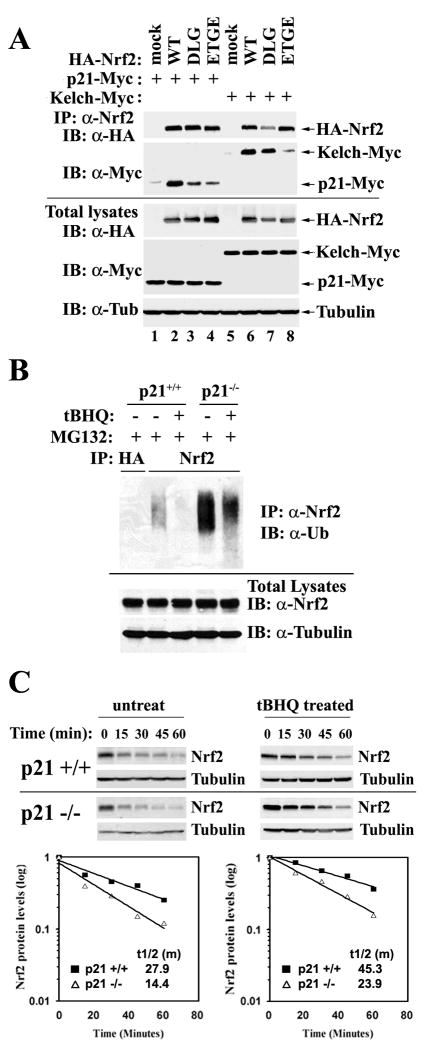
Figure 5. p21 Interfered with the Keap1-Dependent Ubiquitination of Nrf2 by Competing with Keap1 for Nrf2 Binding Mainly through the DLG Motif.
(A) p21 interacted with the DLG and ETGE motifs of Nrf2. Two Nrf2 mutants were generated: mDLG and mETGE in which the DLG or ETGE motif was replaced with alanine residues respectively. COS-1 cells were either cotransfected with expression vectors for the indicated Nrf2 protein and p21-Myc; or expression vectors for the indicated Nrf2 protein and Kelch-Myc. Transfected cells were immunoprecipitated with an anti-Nrf2 antibody and the immunoprecipitated proteins were subjected to immunoblot analysis with anti-HA and anti-Myc antibodies (upper panels). An aliquot of total lysate was analyzed with anti-HA, anti-Myc, and anti-tubulin antibodies (lower panels). (B) p21 reduced ubiquitination of Nrf2. HCT116-p21+/+ and HCT116-p21-/- cells were either untreated or treated with 100 μM tBHQ for 4 hrs in the presence of 10 μM of MG132, which blocks degradation of the ubiquitinated proteins. Cell lysates were subjected to an in vivo ubiquitination assay for detection of the ubiquitin-conjugated endogenous Nrf2 protein. Lysates were denatured and immunoprecipitated with an anti-Nrf2 or an anti-HA antibody (negative control), and blotted with an anti-ubiquitin antibody (upper panel). An aliquot of total lysate was analyzed for Nrf2 and GAPDH expression (lower panels). (C) p21 stabilized Nrf2 in basal and stressed conditions. HCT116-p21+/+ or HCT116-p21-/- cells were left untreated or pre-treated with 100 μm tBHQ for 4 h, followed by addition of 25 μM CHX and incubated for the time periods indicated. Endogenous Nrf2 was detected and the intensity of the Nrf2 bands was quantified and plotted on a semi-log graph. The amount of Nrf2 before addition of CHX was set as 1.