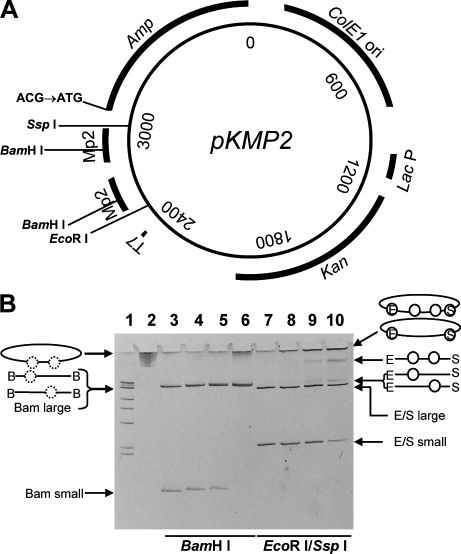
Figure 1.
The pKMP2 plasmid and its charging with nucleosomes. (A) Mutation substrate pKMP2 was used to determine AID-induced cytosine deaminations in the Ampr gene. T7, T7 RNA polymerase promoter for transcription in vitro; MP2, nucleosome positioning sequences; Amp, Ampr gene coding region whose initiation codon ATG has been changed to ACG; ColE1 ori, LacP, and Kan, origin of replication, Lac promoter, and coding region of the Kanr gene for expression of kanamycin resistance in E. coli transformed with pKMP2. Restriction sites used in this study are indicated. (B) Positioned nucleosomes formed on supercoiled pKMP2 plasmid, naked plasmid DNA, and nucleosome containing plasmids with and without restriction enzyme digestion were electrophoresed on a 5% polyacrylamide gel. Lane 1 shows size markers of 1-kb DNA ladder with 500 bp, 517 bp, 1 kbp, 1.5 kbp, 2 kbp, 3 kbp, and 4 kb resolved on the gel. The final band contains four sizes: 5-, 6-, 8-, and 10-kbp DNA molecules. Lanes 2 and 3 show pKMP2 naked DNA uncut or cut with BamHI. Lanes 4–6 show pKMP2 nucleosomes cut with BamHI (0.8, 1.5, and 3.1 µg of histone octamers per 10 µg of DNA, respectively). Histones were removed after BamHI treatment. Lanes 7–10 show pKMP2 naked DNA or nucleosomes (as in lanes 4–6, but histones were not removed after restriction enzyme digestion) cut with EcoRI and SspI. The conformation of the plasmids without and with nucleosomes (circles) are shown. On the left, images for lanes 4–6 are shown. Dotted circles indicate positioned nucleosomes that were removed by removal of histones after BamHI (B) digestion. On the right, images for lanes 8–10 are shown. Circles indicate nucleosomes in EcoRI (E)- and SspI (S)-digested plasmids. The data in B were repeated five times with similar results.