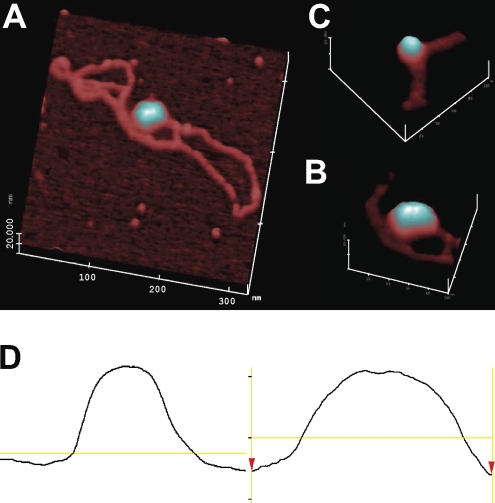
Figure 3.
AFM of pKMP2 plasmids with nucleosomes. (A) Full-length plasmid (DNA contour length measured to be ∼1.1 µm or ∼3.2 kB). The magnification is 200,000×, with a 20-nm full-scale z axis. Using the AFM offline software, the full contour length of the free DNA was segmented into ∼50 linear segments, each ∼20 nm in length, and the segment lengths were tallied. (B) Portion of another pKMP2 plasmid with a single nucleosome. The magnification is 300,000×, with a 10-nm full-scale z axis. (C) The magnification is 300,000×, with a 10-nm full-scale z axis of a portion of the plasmid shown in A. (D) Single-line topographic AFM traces of the mononucleosome (left) and dinucleosome (right) as shown in B and C, respectively. The trace of the dinucleosome shows a bipartite topography and is roughly twice the size of the mononucleosome. The experiment was done twice with similar results.