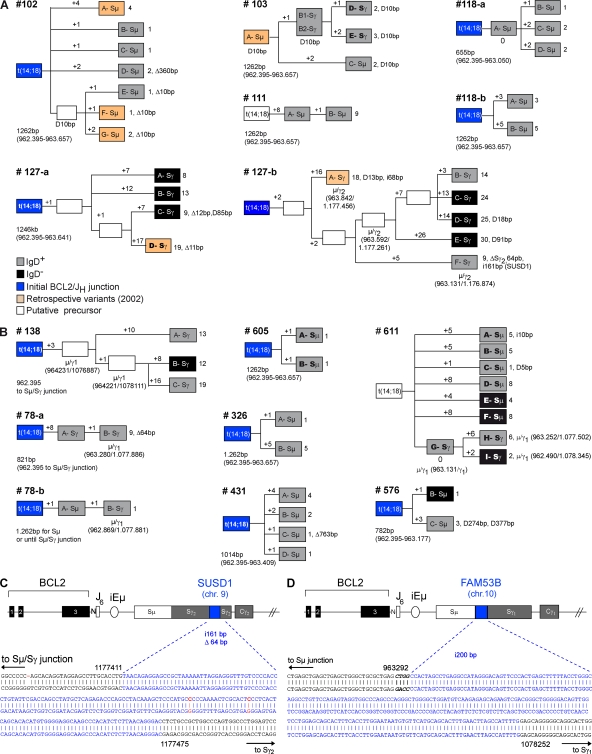
Figure 4.
AID mediates the evolution of t(14;18)+ activated clones in HI. (A and B) Genealogic trees generated from healthy controls (n = 5) and exposed individuals (n = 7) for which several molecular subclones derived from the same BCL2/JH junction (labeled A to G in boxes) were obtained. Trees are organized based on ICV, including SHM in the Sµ region and CSR on both the expressed (gray boxes, sIgD+; black boxes, sIgD−) and the translocated (indicated as Sµ or Sγ in boxes) alleles. For most trees, the BCL2/JH junction was already identified at enrollment (blue boxes, ICV information not available). Six LR-PCR amplicons were obtained from the retrospective PBMC samples (orange boxes) and could be branched in four distinct trees. Stepwise accumulation of mutations is indicated by numbers above the branches (+1 to +26). Total mutations, insertions (i), duplications (D), and deletions (▵) are summarized at the end of each branch. Shared mutations were used to define putative intermediate or precursor cells (open boxes). The length of the analyzed Sµ region is indicated below each tree and varied slightly in case of CSR. (C) Sequence analysis of an AID-mediated foreign DNA insertion (200 bp from the FAM53B gene, chr 10, in blue) into an Sµ/Sγ switch junction of an activated t(14;18)+ clone from sample #138. (D) Sequence analysis of a foreign DNA insertion (161 bp from the SUSD1 gene, chr 9, in blue) into an intra-Sγ2 deletion (▵64 bp) from an activated t(14;18)+ clone from sample #127-b (see clone F-Sγ in A). Numbers refer to germline IGH available from GenBank/EMBL/DDBJ under accession no. NG_001019.