Abstract
Artemis was initially discovered as the gene inactivated in human radiosensitive T−B− severe combined immunodeficiency, a syndrome characterized by the absence of B and T lymphocytes and cellular hypersensitivity to ionizing radiation. Hypomorphic Artemis alleles have also been identified in patients and are associated with combined immunodeficiencies of varying severity. We examine the molecular mechanisms underlying a syndrome of partial immunodeficiency caused by a hypomorphic Artemis allele using the mouse as a model system. This mutation, P70, leads to premature translation termination that deletes a large portion of a nonconserved C terminus. We find that homozygous Artemis-P70 mice exhibit reduced numbers of B and T lymphocytes, thereby recapitulating the patient phenotypes. The hypomorphic mutation results in impaired end processing during the lymphoid-specific DNA rearrangement known as V(D)J recombination, defective double-strand break repair, and increased chromosomal instability. Biochemical analyses reveal that the Artemis-P70 mutant protein interacts with the DNA-dependent protein kinase catalytic subunit and retains significant, albeit reduced, exo- and endonuclease activities but does not undergo phosphorylation. Together, our findings indicate that the Artemis C terminus has critical in vivo functions in ensuring efficient V(D)J rearrangements and maintaining genome integrity.
B and T lymphocytes, the primary cells of the adaptive immune system, provide a major line of defense against foreign molecules by virtue of vastly diverse antigen-specific receptors. The variable regions of antigen receptor genes are assembled from numerous V (variable), D (diversity), and J (joining) coding segments, which are cut and pasted together in a multitude of combinations via the programmed DNA rearrangement V(D)J recombination (1, 2). This process occurs during early lymphocyte development and is required for progenitors to mature into functional B and T lymphocytes. V(D)J recombination is initiated by the recombination activating gene (RAG) 1 and 2 proteins, which comprise an endonuclease that recognizes specific recombination signal sequences (RSSs) adjacent to each V, D, and J segment. The RAG1/2 endonuclease induces DNA double-strand breaks (DSBs) between the RSSs and coding segments (2–4) and generates ends with two distinct structures: 5′ phosphorylated blunt RS ends and covalently closed hairpin coding ends. Joining of the RAG1/2-generated DNA breaks requires the ubiquitously expressed nonhomologous end-joining (NHEJ) DNA repair factors, which include Ku70, Ku80, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4, DNA ligase IV (Lig4), Cernunnos/XLF, and Artemis (1, 2, 5, 6). The NHEJ factors also play critical roles in general DSB repair and maintaining genome stability.
Artemis possesses intrinsic 5′ to 3′ single-strand exonuclease activity and becomes activated as an endonuclease when complexed with DNA-PKcs (7). Artemis–DNA-PKcs nicks the covalently closed coding ends, and hairpin opening at variable positions away from the apex, followed by fill-in synthesis, leads to addition of palindromic “P” nucleotides. Together, these end-processing steps further diversify the antigen receptor genes that are generated upon ligation of modified coding ends. In contrast, the majority of RS ends are precisely joined with no loss or gain of nucleotides; however, the blunt ends can also undergo deletions and N insertions (8–12).
Artemis was initially discovered as the gene mutated in human radiosensitive T−B− severe combined immunodeficiency (RS-SCID), a disorder characterized by a lack of T and B lymphocytes and increased cellular radiosensitivity (13, 14). The majority of Artemis mutations that cause RS-SCID are located within a highly conserved metallo-β-lactamase/β-CASP catalytic domain (residues 1–385 of 692 aa), which distinguishes this family of proteins (14). These mutations include genomic exon deletions, nucleotide deletions, and nonsense mutations and are presumed to inactivate protein function. Inherited hypomorphic Artemis alleles have also been identified in patients and cause combined immunodeficiency syndromes of varying severity, including Omenn syndrome (14–17). Intriguingly, premature translation termination mutations that result in loss of coding sequence within the nonconserved C terminus (aa 386–692) can lead to either RS-SCID (17, 18) or partial immunodeficiency, in which patients have low but detectable B and T lymphocyte populations (15). In this regard, patients harboring a truncating Artemis mutation (D451fsX10, referred to as P70 herein) are characterized by a syndrome of partial immunodeficiency and aggressive EBV-associated lymphoma (15).
The predicted protein encoded by the Artemis-P70 allele retains the metallo-β-lactamase/β-CASP catalytic core of Artemis yet lacks the majority of the nonconserved C terminus (15). The C-terminal domain interacts with and is phosphorylated by DNA-PKcs (19, 20); however, although Artemis and DNA-PKcs form a stable complex in cells, the functional importance of complex formation or Artemis phosphorylation in vivo is not well understood. In this regard, although first proposed to be required for regulation of intrinsic nuclease activities (7), biochemical studies of mutant Artemis proteins have provided evidence that phosphorylation by DNA-PKcs is not necessary for activation of endonucleolytic activities (21). In addition, in vitro cellular assays examining the V(D)J recombination and DNA repair activities of exogenously expressed C-terminally truncated Artemis proteins that lack DNA-PKcs phosphorylation sites, or mutant forms that cannot bind DNA-PKcs, have led to differing results regarding the functional importance of this domain (17, 20, 22, 23). Thus, the in vivo relevance of DNA-PKcs interaction with and/or phosphorylation of the Artemis C terminus remains an open question.
To gain better understanding of the in vivo consequences of the Artemis-P70 hypomorphic allele and to elucidate the functional importance of the C-terminal domain, we generated a mutant mouse strain that models this mutation in the germline via gene targeting. We find that Artemis-P70 homozygous mice exhibit impairment but not a complete block in T and B lymphocyte development, thereby recapitulating the partial immunodeficiency phenotype observed in the patients. We further characterize the impact of this mutation on DNA end processing and V(D)J recombination using combined molecular, cellular, and biochemical approaches. The results of our studies provide insights into the mechanisms that underlie partial immunodeficiency in patients that harbor this hypomorphic allele. In addition, our findings reveal important in vivo functions of the Artemis C terminus in general DNA DSB repair and maintenance of genomic stability.
RESULTS
Development of a mouse model of human partial immunodeficiency
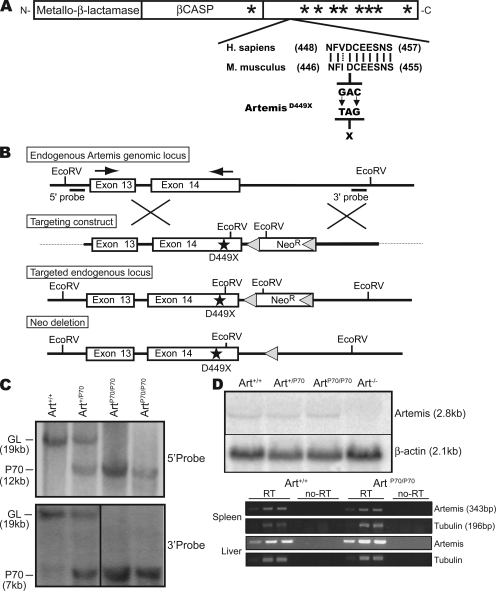
We generated a mouse model harboring the hypomorphic Artemis-P70 allele within the endogenous locus via targeted mutation. The P70 allele is a 7-nt deletion that results in a frameshift at D451 (D449 of the mouse Artemis protein) followed by a premature stop codon 10 aa downstream (15). To model the P70 mutation within the mouse genomic locus, we introduced a nonsense mutation to substitute a stop codon for the aspartate at position 449 (of 603 aa) via site-directed mutagenesis (details in supplemental text). The human and mouse Artemis complementary DNA (cDNA) sequences share ∼73% sequence identity and 81% sequence similarity, and the amino acid sequence of mouse Artemis at the site of the mutation is nearly identical to that of the human protein (Fig. 1 A).
Figure 1.
Generation of Artemis-P70 knock-in mice. (A) Hypomorphic Artemis-P70 mutation. Diagram of Artemis cDNA depicting highly conserved metallo-β-lactamase/β-CASP domain (boxes) and nonconserved C terminus with the approximate positions of DNA-PKcs S/Q phosphorylation sites indicated (asterisks). An alignment between the Homo sapiens and Mus musculus amino acid sequences adjacent to the nonsense mutation introduced into the mouse genomic locus is shown. Lines, identical residues; dotted line, conserved residue; arrows, RT-PCR primers. (B) Targeting strategy for the Artemis-P70 knock-in mutation. The endogenous Artemis locus, targeting construct, knock-in allele, and Neo-deleted knock-in allele are depicted. Bars, 5′ and 3′ probes used to screen ES cell clones; stars, Art-P70 mutation. (C) Southern blot analysis of targeted locus. Southern blot analysis of EcoRV-digested genomic DNA from Art+/+, Art+/P70, and ArtP70/P70 kidneys from Neo-deleted mice, hybridized with the 5′ and 3′ probes. Positions of the germline (GL) and targeted (P70) alleles are indicated. (D) Northern blot and RT-PCR analyses of Artemis-P70 mRNA. 20 µg of total RNA isolated from Art+/+, Art+/P70, ArtP70/P70, and Art−/− MEFs was analyzed by Northern blotting using a probe that hybridizes to the 3′ end of the mouse Artemis mRNA. β-Actin was used as a normalization control. RT-PCR was performed on threefold serial dilutions of total RNA from spleen and liver using primers designed to detect transcripts encoding exons 13–14 and a normalization control (tubulin). Each experiment was repeated at least three times with RNA isolated from two independent cell lines and tissues from two mice of each genotype. Representative results are shown. Black lines indicate that intervening lanes have been spliced out.
The targeting construct was comprised of a 2.5-kbp 5′ homology arm that contained the nonsense mutation in exon 14 and a 2.7-kbp 3′ homology arm (Fig. 1 B). The presence of the nucleotide substitutions (G1388T/C1390G within the coding sequence) and integrity of both homology arms were confirmed by DNA sequence analysis. Homologously targeted recombinants were identified by Southern blotting, and two independently derived subclones of successfully targeted embryonic stem (ES) cell lines were injected into C57BL/6 blastocysts. Germline Art+/P70-Neo mice were subsequently bred to transgenic mice expressing the Cre recombinase from the EIIA promoter to delete the LoxP-flanked Neomycin resistance (Neor) gene during early embryogenesis. Progeny harboring the Neor-deleted mutant Artemis allele were then intercrossed to obtain Art+/+, Art+/P70, and ArtP70/P70 mice (Fig. 1 C). Homozygous Artemis-P70 mice were born in Mendelian numbers (Table S1) and survived into adulthood with no obvious developmental defects.
Expression of the P70 mutant allele was verified by Northern blotting of total RNA isolated from WT, heterozygous, and homozygous Artemis-P70 embryonic fibroblasts with a probe that hybridizes near the 3′ end of the gene (Fig. 1 D). In addition, we performed semiquantitative RT-PCR analyses of Artemis-P70 expression in spleen and liver using gene-specific primers that detect transcripts containing exons 13–14 (Fig. 1 D). The levels of Artemis messenger RNA (mRNA) expressed in the ArtP70/P70 tissues and cells were equivalent to controls as measured by both assays. Thus, the nonsense mutation did not significantly affect the stability of the Artemis mRNA. The RT-PCR products were subcloned and sequenced to verify that the translation stop had been introduced into the proper position (D449X) within the Artemis coding region (unpublished data).
Partial combined immunodeficiency caused by the hypomorphic Artemis-P70 mutation
Patients harboring the hypomorphic Artemis-P70 allele exhibited lymphocytopenia and died of either recurrent infections or lymphoma progression (15). However, in contrast with the T−B−SCID syndrome caused by null mutations in Artemis, low levels of T and B lymphocytes were detected. We examined homozygous Artemis-P70 mice to determine whether the mouse model recapitulated the partial immunodeficiency observed in patients. We observed that the number of thymocytes in ArtP70/P70 mice was drastically reduced compared with WT and heterozygous littermates (10–15% of controls; Table I). In addition, significantly lower numbers of splenocytes were observed in the Artemis-P70 mutant mice, reflecting a decrease in numbers of mature lymphocytes. Importantly, the impact of the hypomorphic Artemis mutation was less severe compared with the inactivating mutation. In this regard, the numbers of thymocytes and splenocytes in ArtP70/P70 mice were notably higher in comparison with Artemis-null mice (Table I).
Table I.
Impact of the Artemis-P70 mutation on lymphocyte development
| Genotype | Thymocytes | CD4+CD8+ thymocytes | Splenocytes | CD4+ LN T cells | CD8+ LN T cells | IgM+ splenocytes | Pro–B cells | Pro–B cells | Pre–B cells | Pre–B cells |
| ×106 | % | ×106 | ×106 | ×106 | % | % | ×105 | % | ×105 | |
| Art+/+ (n = 7) | 167 ± 102 | 75 ± 5 | 70.1 ± 27.95 | 3.57 ± 2.06 | 1.72 ± 0.91 | 19.84 ± 3.82 | 2.38 ± 0.65 | 0.71 ± 0.18 | 15.97 ± 5.69 | 4.63 ± 1.62 |
| Art+/P70 (n = 7) | 220 ± 106 | 78 ± 4 | 94.3 ± 43.2 | 4.61 ± 2.1 | 2.1 ± 0.6 | 18.11 ± 6.2 | 2.37 ± 0.31 | 0.897 ± 0.44 | 10.36 ± 3.25 | 4.34 ± 4.04 |
| ArtP70/P70 (n = 6) | 19 ± 10 | 22 ± 15 | 31.4 ± 19.5 | 0.43 ± 0.25 | 0.59 ± 0.2 | 3.74 ± 1.9 | 4.44 ± 1.79 | 1.34 ± 0.56 | 1.96 ± 0.56 | 0.75 ± 0.33 |
| Art−/− (n = 6) | 3 ± 2 | 7 ± 10 | 15.06 ± 11.4 | 0.059 ± 0.04 | 0.02 ± 0.013 | 0.25 ± 0.21 | 1.5 ± 1.5 | 0.55 ± 0.74 | 0.292 ± 0.23 | 0.1 ± 0.1 |
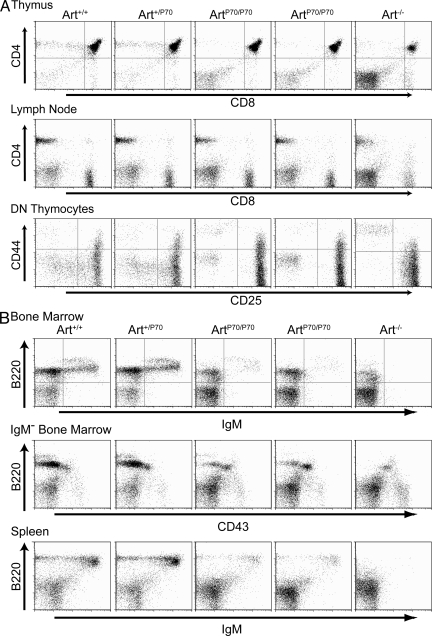
We next performed flow cytometric analyses of cell surface markers present on the lymphocytes to determine the effect of the Artemis-P70 mutation on lymphocyte development. Within the TCR-β locus, Dβ to Jβ rearrangements are initiated as CD4−CD8− (double negative [DN]) thymocytes transition from the CD44+CD25+ DN2 to the CD44−CD25+ DN3 stage of development. Vβ to DJβ rearrangements are completed within the DN3 stage of development, and productive TCR-β rearrangement induces cellular proliferation and differentiation to the CD44−CD25− DN4 stage. The ArtP70/P70 mutant mice exhibited an impairment in T cell development at the DN3 stage, as indicated by an accumulation of progenitors at this stage (Fig. 2 A), which is consistent with a defect in successful completion of TCR-β rearrangement. Despite this defect, a substantial number of mutant developing T cells did progress to the CD4+CD8+ (double positive [DP]) stage in the thymus and CD4+ or CD8+ single-positive stages in the periphery (i.e., lymph nodes and spleen; Fig. 2 A and Table I). In comparison, a more severe block in T cell development was caused by the Artemis-null mutation as a significantly smaller population of thymocytes progressed to the DP stage in a subset of mice (Fig. 2 A and Table I) (24).
Figure 2.
Homozygous Artemis-P70 mice exhibit partial immunodeficiency. Flow cytometric analyses were performed on Art+/+, Art+/P70, ArtP70/P70, and Art−/− mice, as indicated, at 4–5 wk of age. (A) Analysis of T cell development. Thymocytes and lymph node cells were stained with antibodies against the indicated cell surface markers. DN thymocytes were gated for the CD25/CD44 analyses. (B) Analysis of B cell development. Bone marrow and splenocytes were stained with antibodies against the indicated cell surface markers. IgM-negative bone marrow cells (IgM−) were gated for B220/CD43 analyses. Five to six mice of each genotype were analyzed. Representative FACS plots are shown.
Flow cytometric analysis of B lymphocyte development in ArtP70/P70 mice revealed a similar impairment at the progenitor stages when V(D)J rearrangements occur. Within the Ig heavy chain (IgH) locus, DH to JH rearrangements are initiated at the B220+CD43+ pro–B stage, and upon successful VH to DJH rearrangement, cells progress to the B220+CD43− pre–B cell stage. The homozygous Artemis-P70 mice exhibited a defect in the ability of progenitors to transition from the pro–B to pre–B stage of development and, thus, we observed a substantial decrease in the percentage and number of pre–B cells compared with controls (Fig. 2 B and Table I). This impairment in B cell development resulted in significantly lower numbers of surface IgM-expressing cells in ArtP70/P70 bone marrow as well as peripheral lymphoid organs (Fig. 2 B and Table I). In contrast, B cell development was completely blocked at the pro–B stage, and IgM-positive cells were not detected in the bone marrow or periphery in Artemis-null mice (Fig. 2 B and Table I) (24). No measurable lymphocyte development phenotypes were detected in Artemis-P70 heterozygous mice compared with controls. Thus, the immune system phenotypes observed in the Artemis-P70 mutant mice closely parallel the partial immunodeficiency syndrome observed in human patients and are distinct from Artemis nullizygosity.
Defective V(D)J rearrangements in Artemis-P70 homozygous mice
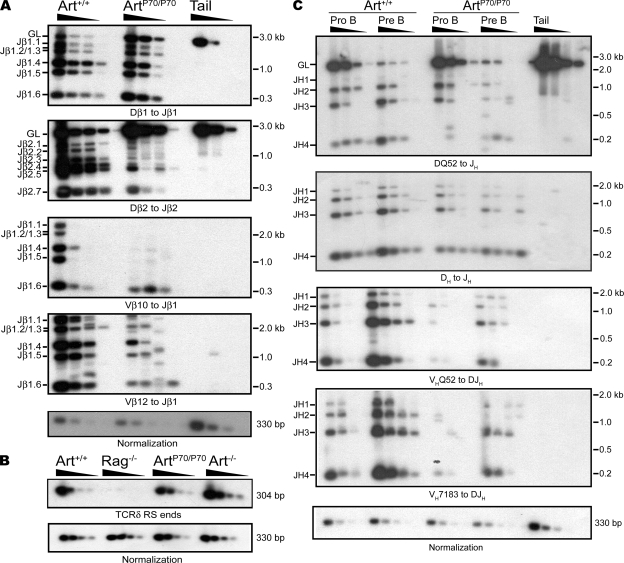
We next examined the impact of the P70 mutation on levels of D to J and V to DJ rearrangements within the IgH and TCR-β loci. To this end, we performed semiquantitative PCR amplification of the rearranging loci followed by Southern blotting in progenitor lymphocyte populations. Within the TCR-β locus, we observed a significant retention of the PCR product corresponding to unrearranged alleles in sorted DN thymocytes isolated from Artemis-P70 homozygous mice compared with controls (Fig. 3 A, GL). We also observed decreased, but readily detectable, levels of PCR products representing Dβ1 to Jβ1 and Dβ2 to Jβ2 rearrangements in ArtP70/P70 DN thymocytes (Fig. 3 A). The levels of Vβ to DJβ1 rearrangements using Vβ10- and Vβ12-specific PCR primers were significantly reduced in the homozygous mutant thymocytes compared with controls (Fig. 3 A). Together, these results indicate that the Artemis-P70 hypomorphic mutation decreases the frequency of both D to J and V to DJ rearrangements within the TCR-β locus, yet substantial levels of recombination do occur.
Figure 3.
Lymphocyte development defects are caused by impaired V(D)J rearrangements. (A) TCR-β rearrangements. Genomic DNA from purified DN thymocytes from Art+/+ and ArtP70/P70 mice and a nonrearranging tissue (tail) was PCR amplified to detect the indicated Dβ to Jβ and Vβ to DJβ rearrangements. (B) Endogenous TCR-δ signal joints. Levels of extrachromosomal signal joints formed between TCR-Dδ2 and -Jδ1 RSSs were detected by PCR amplification of genomic thymocyte DNA isolated from Art+/+, ArtP70/P70, Art−/−, and RAG2−/− mice. Control PCR amplification of a nonrearranging locus was performed to normalize levels of input DNA. Dδ2-Jδ1 signal joints were detected by Southern blot hybridization using an oligonucleotide probe. (C) IgH rearrangements. Genomic DNA from sorted pro- and pre–B cell populations from Art+/+ and ArtP70/P70 mice and a nonrearranging tissue (tail) was PCR amplified to detect DHQ52 to JH and other DH to JH rearrangements using a degenerate DH primer, as well as VH to DJH rearrangements, as indicated. Experiments in A–C were repeated three times with genomic DNA samples from two different sets of mice. Representative results are shown.
We also analyzed DH to JH rearrangements in sorted pro- and pre–B cells from ArtP70/P70 and control mice using the same PCR amplification approach. We used primers 5′ of DHQ52, the D segment located most proximal to JH1, or a degenerate DH primer that amplifies most DH to JH rearrangements, paired with a primer 3′ of JH4. We observed a modest but reproducible decrease in levels of most DH to JH rearrangements in Artemis-P70 pro- and pre–B cells (Fig. 3 C); however, significant levels of DH-JH rearrangements were detected. VH to DJH rearrangements involving two frequently used VH families, VHQ52 and VH7183, were substantially reduced, but not abrogated, in ArtP70/P70 developing B cells (Fig. 3 C). These results indicate that the partial B and T immunodeficiency caused by Artemis-P70 is the result of an impairment, but not complete block, in endogenous V(D)J recombination. Thus, despite loss of approximately one third of the C-terminal region of the protein, the mutant Artemis nuclease possesses the necessary activities required to support in vivo V(D)J recombination, but it is less efficient compared with the WT protein.
DNA end processing during endogenous V(D)J rearrangements in Artemis-P70 mice
To gain a more mechanistic understanding of the V(D)J recombination defects caused by the Artemis-P70 hypomorphic mutation, we analyzed the junctional sequences of unselected endogenous D to J rearrangements. Rearrangements within the IgH and TCR-β loci were PCR amplified, subcloned, and sequenced. The D-JH and D-Jβ1 coding joints from Artemis-null lymphocytes exhibited large deletions, with >400 nt deleted from some coding flanks (Fig. S1) (24). In addition, several unusually long P-nucleotide additions in D-JH and D-Jβ1 junctional sequences were observed (up to 6 nt). Deletions and P nucleotides within D-JH and D-Jβ1 coding joints were also found in WT lymphocytes; however, the largest deletion observed was 12 nt, and none of the P nucleotide additions was longer than 2 bases (Fig. S1).
The majority of coding joints recovered from ArtP70/P70 mice exhibited characteristics of WT junctions (Fig. S1). Most nucleotide deletions at Artemis-P70 coding flanks were <12 nt, and the majority of the P nucleotide additions were 1 or 2 nt long (Fig. S1). We did observe a small proportion of ArtP70/P70 coding joints with features found in Artemis-null junctions, including larger deletions (4 of 51 unique sequences with deletions ranging from 13 to 391 bases) and P nucleotide additions between 3 and 6 bases (6 of 23 coding flanks with P nucleotides). N nucleotide additions were observed in ArtP70/P70 and Art−/− coding joints at a comparable frequency to that in WT joints. However, we noted that both Artemis mutations led to a higher proportion of D-JH and D-Jβ1 junctional sequences containing N nucleotide additions of >5 bases. We observed that of the WT coding joints with N insertions, 6% contained 6 or more nontemplated bases, a proportion which is similar to that in previous reports (24–27). In comparison, 21% of ArtP70/P70 (P = 0.03, Fisher's exact two-tailed test) and 24% of Art−/− (P = 0.02) junctions with N nucleotides contained insertions of >5 bases (Fig. S1) (this study and reference 24).
In patients harboring the P70 mutation, the junctional sequences of eight expressed TCR-β cDNAs exhibited an absence of N nucleotides (15). The precise molecular bases for the differing observations in our mouse model are unclear; however, one potential influencing factor is that the human subjects experienced multiple recurrent infections. Thus, aberrant immune responses in the context of lymphopenia may have had an impact on the selected TCR repertoire of the low-level mature peripheral T cells observed in the patients (28–30). In our analyses, we have examined junctional sequences of >70 unique D to J rearrangements in immature lymphocyte populations from Artemis mutant mice housed in a specific pathogen-free environment; thus, the mouse sequences may allow for a more unbiased analysis of junctional diversity. In this regard, a recent study reported the sequences of DH-JH coding joints recovered from immature B cells isolated from bone marrow mononuclear cultures of Artemis mutant patients, including one patient harboring a distinct premature translation termination mutation (17). The majority of DH-JH junctional sequences analyzed contained N nucleotides, and one third of the coding joints with N additions contained insertions of >5 bases.
We also examined the levels and fidelity of RS end joining in the Artemis-P70 homozygous mice. RS joints formed upon precise ligation of heptamers can generate the recognition sequence for ApaLI by virtue of the conserved CAC triplet flanking the majority of RAG1/2 cleavage sites (31, 32). A proportion of blunt RS ends undergoes modification in WT lymphocytes and, thus, is resistant to digestion by ApaLI because of N nucleotide additions and/or deletions (8–12). Artemis does not play a direct role in RS end joining; however, it has been implicated in processing the blunt ends before ligation (11). In this regard, levels of RS joint formation are unaffected in Art−/− mice; however, the proportion of ApaLI-resistant junctions at certain loci have been found to be decreased, in part as the result of an absence of nucleotide deletions (11). We assessed the impact of the Artemis-P70 mutation on RS end processing and joining within the TCR-δ locus using a PCR strategy. We amplified the joints formed between the Dδ2 and Jδ1 RSSs and observed that the levels were similar in ArtP70/P70 thymocytes compared with Art+/+ and Art−/− thymocytes (Fig. 3 B). These results indicate that Artemis-P70 does not significantly impact the frequency of RS joining.
We next subcloned the PCR products, and individual clones were digested with ApaLI to determine the proportion of precise and imprecise joints. The majority of Dδ2-Jδ1 RS junctions from WT thymocytes were precisely joined (80%, corrected for redundancy; Table S2), and a comparable proportion (70%) of Art−/− RS joints were sensitive to ApaLI digestion. We found that a significantly lower percentage of ArtP70/P70 RS joints (64%; P = 0.03, Fisher's exact two-tailed test) were precise in comparison with WT junctions (Table S2). Sequence analysis of the imperfect RS joints from WT, Art−/−, and ArtP70/P70 mice revealed that nearly all of the junctions analyzed contained N nucleotide additions (Fig. S2). We also observed deletions from Dδ2 or Jδ1 RS ends in a small subset of RS joints analyzed from WT (1 of 15 unique imprecise junctions) as well as Art−/− (2 of 18) and ArtP70/P70 (2 of 28) thymocytes. Together, these results indicate that the Artemis-P70 mutation has a modest impact on the level of fidelity of Dδ2-Jδ1 RS joining. As the frequency of deleted ends was similar in all genotypes examined, our findings suggest that the decreased levels of precise RS joining in ArtP70/P70 mice is the result of an increase in N nucleotide–modified ends.
The observed nucleotide loss within two Artemis-null Dδ2-Jδ1 RS joints differs from a previous study that found an absence of deletions from Dδ1-Dδ2 and Vαδ-Dδ2 RS junctions in Artemis deficiency (11). These incongruent results may be caused by distinct mechanisms of end processing at the different recombining V, D, and J segments. In this regard, the extent of N nucleotide addition and deletion of RS ends varies according to the sequences of flanking coding segments and the particular loci undergoing rearrangement (8–12). Thus, involvement of Artemis nuclease activities in RS end modification may differ depending on the specific rearranging gene segments. Nonetheless, these findings support the notion that Artemis is not only required for processing hairpin coding ends during V(D)J recombination but also participates in modification of blunt RS ends. Furthermore, our results indicate that the C-terminal portion of the nuclease that is absent in the Artemis-P70 protein plays an important role during these activities.
Analysis of V(D)J recombination intermediates in Artemis-P70 mutant lymphocytes
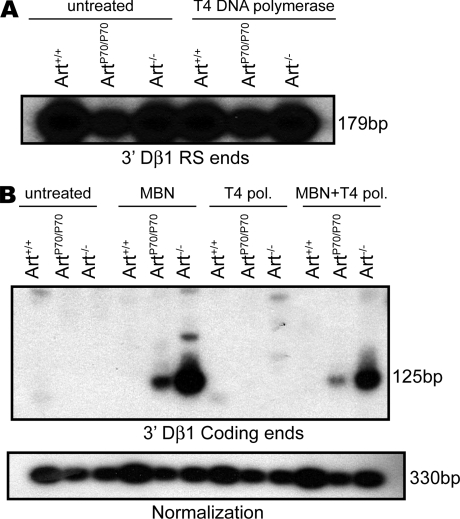
In WT developing lymphocytes, RAG1/2-generated hairpin coding ends are rapidly processed and joined and, thus, do not accumulate to detectable levels (33, 34). In contrast, blunt RS ends persist within the context of a stable postcleavage complex before being ligated (33, 35). Covalently closed coding ends accumulate in Artemis-null lymphocytes as a result of the inability to endonucleolytically cleave the hairpins (24). To assess the impact of Artemis-P70 on end processing, we examined the levels and structures of hairpin coding and RS ends in developing lymphocytes. Genomic DNA isolated from Art+/+, ArtP70/P70, and Art−/− thymocytes was treated with mung bean nuclease (MBN) and/or T4 DNA polymerase. MBN nicks hairpins and can endonucleolytically cleave overhanging sequences, and T4 DNA polymerase can act upon 3′ or 5′ overhanging ends via its intrinsic exonuclease and polymerase activities, thereby rendering them blunt. Thus, enzymatic treatment of DNA ends with either or both enzymes followed by ligation-mediated PCR (LM-PCR) can reveal distinct structures of coding ends.
We ligated linkers to untreated genomic thymocyte DNA or DNA treated with T4 DNA polymerase and then PCR amplified products corresponding to 3′ Dβ1 RS ends within the rearranging TCR-β locus. The LM-PCR products were detected via Southern blot analyses using an oligonucleotide probe. We readily detected RS ends in untreated genomic DNA isolated from ArtP70/P70 thymocytes (Fig. 4 A). To assess the presence of modified nonblunt RS ends, we treated the DNA with T4 polymerase before the linker ligation. We did not observe a significant increase in band intensity or the appearance of additional bands (Fig. 4 A); thus, the majority of Dβ1 3′ RS ends in ArtP70/P70 thymocytes are blunt, as observed in WT and Art−/− mice. Modified RS ends are likely to be present in developing thymocytes, as indicated by the presence of imprecise RS joints containing N nucleotides and deletions; however, accurate assessment of the levels is beyond the sensitivity of the LM-PCR assay (35, 36).
Figure 4.
V(D)J recombination intermediates in Artemis-P70 thymocytes. (A) Levels of RS ends in Artemis-P70 DN thymocytes. Genomic DNA from sorted DN thymocytes from Art+/+, ArtP70/P70, and Art−/− mice was either untreated or treated with T4 DNA polymerase. Linkers were ligated to the blunt ends, followed by PCR amplification of TCR-Dβ1 3′ RS ends (179 bp). The LM-PCR products were detected by Southern blot hybridization using locus-specific oligonucleotide probes. (B) Accumulation of coding hairpin ends in Artemis-P70 DN thymocytes. The genomic DNA from sorted DN thymocytes was untreated or treated with MBN and/or T4 DNA polymerase, as indicated, and then the LM-PCR reactions were performed to detect 3′ TCR-Dβ1 coding ends (125 bp), as described in A. Control PCR amplification of a nonrearranging locus was performed to normalize levels of input DNA. Experiments in A and B were repeated a minimum of three times with genomic DNA isolated from at least two different mice of each genotype. Representative results are shown.
We next assessed the impact of the Artemis-P70 mutation on levels of Dβ1 3′ coding ends in DNA samples treated with MBN and/or T4 polymerase. We detected substantial levels of LM-PCR products corresponding to hairpin coding ends in ArtP70/P70 thymocytes; however, the levels were consistently three- to fivefold lower than those observed in Artemis-null cells (Fig. 4 B). Similar results were observed at Jβ1 5′ coding ends (unpublished data). Hairpin coding ends were not detected in WT thymocytes, as has been previously established (33, 34). Treatment of the thymocyte DNA samples with T4 DNA polymerase did not impact the levels of LM-PCR products in any of the genotypes (Fig. 4 B); thus, open hairpins with 5′ or 3′ overhanging ends do not persist in ArtP70/P70 thymocytes. These findings support the notion that the Artemis-P70 mutant protein is defective for hairpin coding end opening; however, it does possess moderate in vivo endonucleolytic activity, as indicated by reduced accumulation of coding ends compared with Artemis nullizygosity.
Coding joining defects in Artemis-P70 mutant cells
To gain additional mechanistic insights into the V(D)J recombination defects caused by the Artemis-P70 mutation, we used a plasmid-based assay in cultured cells (37). Initially, we analyzed the ability of mutated human Artemis cDNA expression constructs to complement the V(D)J recombination defects in Artemis-null ES cells (38). We generated three C-terminal truncations, including D451X, which models the human P70 mutation, T432X, and S385X via site-directed mutagenesis. The T432X protein mimics a distinct human Artemis hypomorphic frameshift mutation that is associated with partial immunodeficiency (15). The S385X C-terminally truncated protein is comprised of the highly conserved metallo-β-lactamase/β-CASP domain that lacks the entire nonconserved C terminus and represents the catalytic core (23). The mutant or WT Artemis constructs were cotransfected into Art−/− ES cells with RAG1 and RAG2 expression constructs and a coding joining plasmid substrate (pJH290). Levels of V(D)J recombination were quantitated by transformation of recovered plasmids into Escherichia coli and selection of successfully recombined products via ampicillin and chloramphenicol resistance. We observed that all mutant forms of Artemis complemented the severe coding joining defect (Table II).
Table II.
C-terminally truncated Artemis proteins complement Artemis-null V(D)J coding joining defect
| Cell lines | AmpR/AmpR + ChlR | Recombination frequency | Relative levels |
| % | |||
| Experiment 1 | |||
| TC1 | 330/24,000 | 1.3 | 1.0 |
| Art−/− | 35/20,100 | 0.17 | 0.13 |
| Art−/− + hArt | 3,410/250,000 | 1.4 | 1.08 |
| Art−/− + D451X | 840/42,500 | 2 | 1.54 |
| Art−/− + T432X | 5,500/405,000 | 1.3 | 1.0 |
| Art−/− + S385X | 460/42,000 | 1.09 | 0.84 |
| TC1 (no RAG) | 0/2,600 | <0.038 | <0.029 |
| Experiment 2 | |||
| TC1 | 1,753/285,066 | 0.615 | 1.0 |
| Art−/− | 278/99,467 | 0.279 | 0.45 |
| Art−/− + hArt | 5,370/194,867 | 2.76 | 4.5 |
| Art−/− + D451X | 2,045/82,133 | 2.49 | 4.1 |
| Art−/− + T432X | 3,819/178,000 | 2.15 | 3.5 |
| Art−/− + S385X | 1,444/103,133 | 1.40 | 2.3 |
| TC1 (no RAG) | 73/50,800 | 0.144 | 0.2 |
We analyzed the junctional sequences of recombined plasmids to identify qualitative differences in coding end processing and found that coding joint sequences recovered from Art−/− cells expressing the mutant Artemis constructs were similar to those from WT ES cells or mutant cells complemented with WT Artemis (Fig. S3). Overall, the coding joint sequences exhibited similar numbers of P and N nucleotide additions and deletions. The rare recombinants isolated from Artemis-null cells exhibited the hallmark large deletions and longer than average P nucleotide additions. These results demonstrate that the hypomorphic Artemis mutations that lead to partial immunodeficiency in patients, D451X and T432X, retain the necessary catalytic activities to properly process the hairpin coding ends during V(D)J recombination when overexpressed. These findings are in agreement with previous studies, which reported that the DNA-PKcs interaction domain of Artemis was largely dispensable for V(D)J recombination in cells using model substrates (17, 20, 22, 23). However, these in vitro results did not recapitulate our in vivo observations of impaired V(D)J rearrangements and hairpin coding end accumulation within the endogenous antigen receptor loci in ArtP70/P70 lymphocytes.
To more accurately assess the impact of the Artemis-P70 mutation on V(D)J recombination, we compared frequencies of coding joint formation in Art+/+, Art+/P70, ArtP70/P70, and Art−/− primary transformed mouse embryonic fibroblasts (MEFs). We observed a dramatic decrease in recombination frequency in Artemis-P70 homozygous cells, which was of a similar magnitude to that observed in Artemis-null MEFs (Table III). Coding joining in ArtP70/P70 cells was restored nearly to WT levels by expression of the full-length Artemis protein. These results in cells expressing endogenous levels of the Artemis-P70 protein are in sharp contrast to the complementation assay described in the previous section and emphasize the potential limitations of heterologous overexpression of mutant proteins in characterizing physiological activity. Conflicting reports concerning the V(D)J recombination activities of mutant Artemis proteins are likely influenced by variable levels of exogenous expression (17, 20, 22, 23).
Table III.
Defective V(D)J coding joining in homozygous Artemis-P70 MEFs
| Cell lines | AmpR/AmpR + ChlR | Recombination frequency | Relative levels |
| % | |||
| Experiment 1 | |||
| WT (No Rag) | 2/9,400 | 0.02 | 0.02 |
| WT | 399/36,500 | 1.09 | 1.0 |
| Art+/P70 | 2,514/84,800 | 2.96 | 2.7 |
| ArtP70/P70 | 7/51,100 | 0.01 | 0.01 |
| Art−/− | 17/30,900 | 0.05 | 0.05 |
| Experiment 2 | |||
| WT (No Rag) | 5/23,800 | 0.02 | 0.06 |
| WT | 48/9,800 | 0.49 | 1.0 |
| Art+/P70 | 104/15,400 | 0.68 | 1.4 |
| ArtP70/P70 | 5/50,800 | 0.01 | 0.02 |
| Art−/− | 8/50,800 | 0.02 | 0.03 |
| Experiment 3 | |||
| WT | 1,933/602,500 | 0.32 | 1.00 |
| ArtP70/P70 | 10/51,750 | 0.02 | 0.06 |
| ArtP70/P70 + hArt | 66/29,750 | 0.22 | 0.69 |
| Art−/− | 67/148,000 | 0.03 | 0.08 |
| Art−/− + hArt | 1,462/282,500 | 0.52 | 1.63 |
| No RAG | 0/6,500 | <0.02 | <0.06 |
Qualitative analyses of the coding joints recovered from the ArtP70/P70 MEFs revealed that, despite the severe decrease in recombination frequency, the sequences of the coding joints were similar to those observed in WT MEFs (Fig. S4). The majority of Artemis-P70 junctions were characterized by P and N nucleotide additions of normal length and small nucleotide deletions, with the exception of two junctions exhibiting larger deletions (>12 bases). The joints from Artemis-null MEFs were clearly distinct and most exhibited large deletions from both coding flanks (Fig. S4). Together, these findings support the notion that the Artemis C terminus is not required for intrinsic catalytic activities and indicate that this region of the nuclease facilitates efficient coding end processing and joining.
Hypomorphic Artemis mutation results in intermediate hypersensitivity to DSBs and genome instability
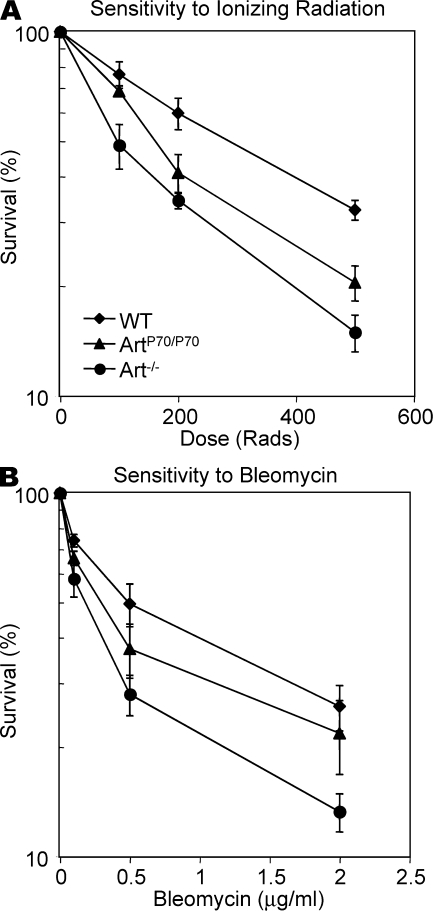
One hallmark feature of cell lines derived from human RS-SCID patients harboring null mutations in Artemis is increased sensitivity to DNA DSB-inducing agents, such as ionizing radiation (IR), and the radiomimetic chemotherapeutic agent bleomycin, and Artemis-null mouse cells recapitulate this phenotype (13, 24, 38). To determine the impact of the Artemis-P70 mutation on general DNA DSB repair, we examined the relative sensitivities of Art+/+, ArtP70/P70, and Art−/− primary MEFs. We found that the ArtP70/P70 MEFs exhibited an intermediate hypersensitivity to IR compared with Artemis nullizygosity, which is consistent with a partial loss of DNA repair functions (Fig. 5 A). We also consistently observed that Artemis-P70 homozygosity resulted in intermediate survival upon exposure to bleomycin, although the impact was less pronounced compared with IR hypersensitivity (Fig. 5 B).
Figure 5.
Defective DNA repair and increased genomic instability caused by the Artemis-P70 mutation. (A) IR sensitivity. Art+/+, ArtP70/P70, and Art−/− primary MEFs were exposed to the indicated amounts of IR and then plated in duplicate. The cells were harvested 7 d after IR, stained with trypan blue, and then counted. The percentage of survival compared with untreated cultures is plotted as a function of IR dose. (B) Bleomycin sensitivity. Primary MEFs were plated and then treated with the indicated amounts of bleomycin at 24 h. Cellular survival was determined as described in A. The curves represent the mean of three independent experiments using two independent cell lines of each genotype. Error bars represent SD.
We next assessed the impact of the Artemis-P70 mutation on chromosomal stability in early passage primary MEFs. Artemis-null cells accumulate increased levels of chromosomal aberrations (24, 38), which likely result from the presence of unrepaired DNA ends. We found that ArtP70/P70 cells harbored increased levels of chromosomal aberrations at 24 h after 200 rads IR, particularly chromosome and chromatid breaks, compared with the WT cells (Table IV). The level of aberrations in Art-P70 MEFs was similar to that observed in Artemis-null cells; thus, the P70 hypomorphic Artemis mutation leads to significantly increased genome instability. These results support the notion that the hypomorphic P70 allele results in partial loss of Artemis function, not only for end processing during the lymphoid-specific V(D)J recombination but also general DSB repair.
Table IV.
Increased IR-induced chromosomal anomalies in hypomorphic Artemis-P70 MEFs
| Type of anomaly | Art+/+ | ArtP70/P70 | Art−/− |
| Chromosome/chromatid breaks | 15 | 73 | 50 |
| Fusions | 7 | 16 | 11 |
| Radial structure | 0 | 4 | 0 |
| Ring structure | 6 | 6 | 13 |
| Unusual structures | 2 | 13 | 18 |
| Total anomalies | 30 | 108 | 92 |
| Chromosomes scored | 2,053 | 2,585 | 1,541 |
| Metaphases scored | 54 | 58 | 51 |
| Number of anomalies per chromosome | 0.015 | 0.042 | 0.060 |
| Number of anomalies per metaphase | 0.56 | 1.86 | 1.80 |
Interaction and phosphorylation of mutant Artemis proteins with DNA-PKcs
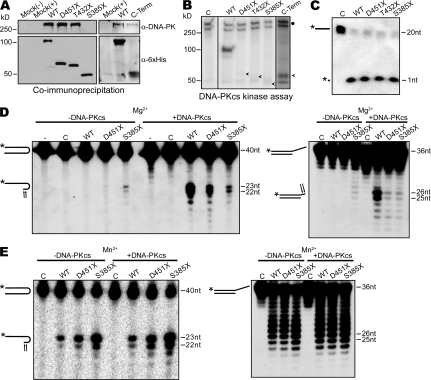
To elucidate the molecular defects caused by the Artemis-P70 mutation, we examined the consequences of this mutation on interactions with DNA-PKcs. Using a coimmunoprecipitation assay, we directly examined the ability of the Artemis-D451X (P70) and the T432X and S385X proteins to interact with DNA-PKcs. Histidine and myc-tagged WT and mutant Artemis proteins were expressed in 293T cells and then immunoprecipitated using an anti-myc antibody. The coimmunoprecipitated proteins were analyzed by Western blotting using anti-histidine and anti–DNA-PKcs monoclonal antibodies. We observed that the D451X and T432X proteins stably interacted with DNA-PKcs to a similar extent as full-length Artemis; however, the S385X C-terminally truncated protein did not (Fig. 6 A). We also expressed the regions of Artemis deleted in D451X and S385X proteins and found that the C terminus (aa 386–692) retains the ability to coimmunoprecipitate DNA-PKcs (Fig. 6 A); however, a polypeptide comprised of residues 452–692 does not (unpublished data). These results are consistent with previous studies that identified the nonconserved Artemis C terminus as important for stable DNA-PKcs binding (20, 22).
Figure 6.
Impact of Artemis C-terminal truncations on DNA-PKcs interactions and nuclease activities. (A) Coimmunoprecipitation of Artemis–DNA-PKcs. Constructs expressing c-myc and 6× histidine-tagged full-length (WT), D451X, T432X, and S385X mutant forms of Artemis, and the C terminus alone (C-term), were transfected into 293T cells, immunoprecipitated with α–c-myc, and then analyzed by Western blotting probed with α-6×His antibodies. The membranes were then reprobed with α–DNA-PKcs antibodies. Mock, untransfected 293T cells; (−), no immunoprecipitation; (+), immunoprecipitated. (B) Phosphorylation of Artemis mutants. WT and mutant Artemis proteins (as indicated) purified from 293T cells were incubated with purified DNA-PKcs and γ-[32P]ATP. Phosphorylation products were analyzed by SDS-PAGE followed by autoradiography. Arrowheads, positions of truncated mutant proteins; closed circle, autophosphorylated DNA-PKcs. Black lines indicate that intervening lanes have been spliced out. (C) 5′ to 3′ single-strand exonuclease activity. WT and mutant Artemis proteins (as indicated) were incubated with a 32P 5′ end-labeled single-strand oligonucleotide substrate (20 nt) for 2 h at 37°C. The reactions were analyzed on a 17% denaturing polyacrylamide gel followed by autoradiography. The positions and structures of the input substrate and 1-nt product are indicated on the left. Asterisks, 32P 5′ end label. (D) DNA-PKcs–dependent endonuclease activities in the presence of MgCl2. Endonuclease reactions were performed with a 32P 5′ end-labeled 20-bp hairpin substrate (left) or a 3′ overhang substrate comprised of a [32P] 5′ end-labeled 36-mer annealed to a complementary 21-mer (right) in the presence or absence DNA-PKcs and ATP, as indicated. All reactions contained 10 mM MgCl2 and were analyzed as described in A. The positions and structures of the input substrates and products are indicated. Arrows, sites of endonucleolytic cleavage; asterisks, 32P 5′ end label. (E) DNA-PKcs–independent hairpin opening activity in the presence of MnCl2. Endonuclease reactions were performed with the 32P 5′ end-labeled hairpin and 3′ overhang substrates as described in B, except, the reactions contained 10 mM MnCl2. Reactions were analyzed as described in A. The positions and structures of the input substrates and products are indicated. C, mock transfection; (−), input substrate. All assays were repeated at least three times with proteins from three or more independent transfections. Asterisks, 32P 5′ end label.
We examined the ability of the mutant Artemis proteins to be phosphorylated by DNA-PKcs, as the majority of phosphorylation sites have been mapped to the nonconserved C-terminal region (19, 21, 22). To this end, we purified the WT, D451X, T432X, and S385X proteins from 293T cells and then incubated them with DNA-PKcs in the presence of γ-[32P]ATP and duplex DNA. We observed a substantial reduction in levels of phosphorylation of D451X, T432X, and S385X compared with WT Artemis, whereas the C terminus alone is efficiently phosphorylated (Fig. 6 B). These results indicate that the protein encoded by the mutant Artemis P70 allele, D451X, is capable of interacting with DNA-PKcs; however, it does not undergo phosphorylation.
Impact of Artemis mutations on intrinsic nuclease activities
We next analyzed the nuclease activities intrinsic to Artemis-D451X and regulation by DNA-PKcs. Initially, we examined the activity of full-length D451X and S385X proteins on a 32P 5′ end-labeled single-strand oligonucleotide substrate and observed that the truncated enzymes retained robust 5′ to 3′ exonuclease activity, as indicated by release of the single labeled nucleotide (Fig. 6 C). We next assayed hairpin opening activity in the presence or absence of DNA-PKcs. In the presence of DNA-PKcs, the full-length protein was activated to cleave the 32P 5′ end-labeled 20-bp hairpin substrate to yield major products of 22 and 23 bases at positions 2 and 3 nt 3′ of the apex (Fig. 6 D, left) (7). We observed that the D451X protein exhibited significant, although reduced, DNA-PKcs–dependent hairpin opening activity, yielding products of the same length as WT Artemis (Fig. 6 D). We also examined the levels of DNA-PKcs–dependent and –independent cleavage activity on a 15-nt 3′ overhang substrate comprised of a 32P 5′ end-labeled 36-mer annealed to a 21-mer. The D451X protein exhibited endonucleolytic activity that was dependent on DNA-PKcs and primarily cleaved the substrate at positions 4 and 5 bases 3′ of the double-to-single strand transition, as observed for WT Artemis (Fig. 6 D, left). Thus, Artemis-D451X possesses DNA-PKcs–dependent endonucleolytic activities on hairpin and 3′ overhang structures; however, we consistently observed lower levels of cleavage products compared with the WT enzyme. The S385X protein exhibited DNA-PKcs–independent endonucleolytic activity on the hairpin and 3′ overhang substrates that was slightly stimulated upon addition of the protein kinase, which is consistent with previous analyses of a similar truncated form of Artemis (aa 1–382) (20). To further evaluate the endonuclease activities intrinsic to the mutant Artemis proteins, we assayed hairpin and 3′ overhang cleavage in the presence of manganese (Mn2+), rather than magnesium (Mg2+), as the divalent metal cofactor. We found that WT Artemis, as well as the D451X and S385X proteins, possesses hairpin nicking activity in Mn2+ which is independent of DNA-PKcs and that this activity is not further stimulated upon addition of the kinase (Fig. 6 E, left). In fact, we observed that in reaction mixtures containing Mn2+, the levels of hairpin cleavage were consistently higher for D451X and S385X than for the full-length protein. The WT, D451X, and S385X proteins also exhibited significant DNA-PKcs independent 3′ overhang cleavage activity in Mn2+ that was not enhanced by the presence of DNA-PKcs (Fig. 6 E, right). We noted that Mn2+ led to the appearance of a range of products corresponding to additional cleavage sites within the 3′ single-strand flap. Thus, the presence of Mn2+ abrogates the requirement for DNA-PKcs in activating endonucleolytic activities of Artemis and circumvents the need for remodeling of the overhang substrate and/or conformation of protein complexes by the protein kinase, as has been proposed for DNA-PKcs–dependent overhang cleavage in Mg2+ (20). Together, these results demonstrate that the C-terminally truncated proteins possess robust endonucleolytic activity in the absence of phosphorylation and, in the case of S385X, without stable interaction with DNA-PKcs under certain reaction conditions.
DISCUSSION
A mouse model of partial immunodeficiency caused by a hypomorphic Artemis mutation
In this study, we examined the molecular mechanisms underlying a syndrome of partial B and T immunodeficiency associated with lymphoma in patients harboring a hypomorphic Artemis allele, P70. The Artemis-P70 mutation results in loss of coding sequence for over one third of the polypeptide, including the region of the C terminus that contains the majority of phosphorylation sites for DNA-PKcs (19, 21, 22). We demonstrate that lymphocyte development in ArtP70/P70 mice is impaired at the stages during which TCR and Ig gene rearrangements occur, thereby resulting in a marked decrease in numbers of mature B and T lymphocytes compared with Art+/P70 and WT controls. However, the phenotype is less severe compared with Art−/− mice. In this regard, ArtP70/P70 mice have substantial populations of CD4+CD8+ DP thymocytes and detectable levels of CD4+ or CD8+ single-positive peripheral T cells as well as B220+IgM+ bone marrow and peripheral B cells. In contrast, Artemis-null mice are characterized by a block in B cell development and only a subset of mice exhibit “leaky” T cell development (this study and reference 24). Thus, the phenotypes of homozygous Artemis-P70 mice are clearly distinct from those observed in Artemis nullizygosity and confirm that the mutation represents a partial loss–of-function allele.
We found that the lymphocyte development defects observed in ArtP70/P70 mice are a result of reduced efficiency of coding end processing during endogenous V(D)J recombination, as indicated by an accumulation of hairpin ends and lower levels of both D to J and V to DJ rearrangements within TCR and Ig loci. We observed a similar defect in V(D)J recombination using a cellular transient transfection V(D)J recombination assay in Artemis-P70 homozygous mutant MEFs. Therefore, although previous in vitro cellular transfection studies examining the functional consequences of Artemis mutations concluded that neither the C terminus nor DNA-PKcs–dependent phosphorylation is required for V(D)J recombination (19, 21–23), our findings clearly establish that the C-terminal region of the protein is indeed required for efficient in vivo V(D)J rearrangements.
Consequences of deletion of the Artemis C terminus on DNA end processing
We found that the majority of coding joint sequences from the ArtP70/P70 mutant mice and cells exhibit similarity to WT junctions. The Artemis-P70 junctions harbored small deletions and P nucleotide additions of normal length, and only a small subset of sequences contained the hallmark long P insertions or large deletions characteristic of Art−/− junctions. These results indicate that the Artemis-P70 mutant protein can be targeted to coding ends and nick the hairpins once recruited, although with reduced efficiency compared with the WT enzyme. We also observed aberrant N nucleotide additions within both coding and RS joints in the ArtP70/P70 mice. In this regard, significantly higher frequencies of DH-JH and Dβ1-Jβ1 coding joints containing N insertions of longer than 5 bases and imprecise RS joints containing N nucleotides were observed. These findings suggest that, in addition to hairpin nicking, Artemis may function to nucleolytically trim TdT-synthesized nontemplated bases added to both coding and RS ends and that loss of the Artemis C terminus has an impact on these activities. Alternatively, Artemis may modulate TdT interactions with and/or accessibility to DNA ends, thereby altering TdT-dependent end processing in the mutant lymphocytes.
Our studies revealed that primary homozygous Artemis-P70 MEFs are hypersensitive to the DNA DSB-inducing agents IR and bleomycin but less sensitive than Art−/− MEFs. Truncation of the Artemis C terminus also results in increased genomic instability, as we observed significantly higher levels of chromosomal anomalies in irradiated ArtP70/P70 MEFs. Therefore, the impact of the P70 mutation on Artemis function is not restricted to end processing during the specialized process of V(D)J recombination but also impacts its roles in general DSB repair and maintenance of genome stability.
What are the consequences of C-terminal truncation on Artemis activities? We found that the D451X protein, which was modeled after the P70 mutant allele, retains robust 5′ to 3′ single-strand exonuclease activity. The truncated protein interacts stably with DNA-PKcs and exhibits significant, although reduced, DNA-PKcs–dependent endonucleolytic activity despite the inability to be phosphorylated. These results are consistent with previous studies indicating that phosphorylation within the Artemis C terminus is dispensable for activation of endonucleolytic activity (21). Our in vitro observations that the V(D)J recombination defect in Artemis-null cells can be fully complemented by exogenous expression of the D451X protein support the notion that the C terminus is not essential for Artemis end processing activities per se. However, we did observe consistently lower levels of endonucleolytic activities intrinsic to the D451X protein, indicating that the C terminus plays a role in modulating Artemis nuclease activities independent of DNA-PKcs interaction. In this regard, the reduced 3′ overhang cleavage activity observed in vitro may result in the distinct N nucleotide additions observed within coding and RS joints in vivo.
One additional in vivo impact of C-terminal truncation and resultant loss of Artemis phosphorylation may be mislocalization or inefficient recruitment of the mutant protein to DNA ends. The phosphorylated form of Artemis forms discrete nuclear foci in cells (22) and preferentially associates with chromatin in response to DNA damage (22, 39), suggesting that phospho-Artemis may have a distinct function within the context of chromatin. Although the precise nature of this function is unclear, cofractionation of phospho-Artemis with DNA-PKcs, XRCC4, and Ku in response to DNA damage suggests that localization to chromatin facilitates repair of DNA DSBs (39). Likewise, in vitro and cellular studies indicate that an autophosphorylated form of DNA-PKcs has a higher affinity for the phosphorylated form of Artemis (unpublished data and references 19, 20). The truncated Artemis-P70 protein lacking the major phosphorylation sites may not properly interact with chromatin or other DNA repair factors. Thus, the hypomorphic mutation would impair the ability of Artemis to be stably targeted to DSBs and have an impact on its ability to efficiently act upon DNA ends.
Implications of aberrant Artemis nucleolytic activities
As aberrant repair of DNA DSBs can lead to oncogenic chromosomal aberrations and cancer (25, 40–42), our findings raise the possibility that the hypomorphic Artemis-P70 allele may predispose to tumorigenesis. In support of this notion, patients harboring the Artemis-P70 mutation developed aggressive B lymphoma (15). The molecular basis for tumorigenesis in these patients was not clear, as the lymphomas were associated with EBV yet also exhibited characteristics that are not generally associated with EBV-driven B cell proliferation, including a clonal cellular origin, increased genome instability, and clonal chromosomal aberrations (15). Our observations that the Artemis-P70 allele results in defective DNA DSB repair and increased genome instability provide evidence that this hypomorphic mutation could contribute to tumorigenesis, as observed in mouse models harboring null alleles in NHEJ genes in the context of p53 mutation (25, 41, 42). In further support of this hypothesis, two relatives of a patient harboring a similar hypomorphic Artemis allele, which leads to a frameshift at codon 393 followed by a stop mutation after 8 codons, were diagnosed with acute lymphoblastic leukemia; however, the patient died of pneumonia at 3 mo of age, before developing overt lymphoid malignancy (18). In future studies, it will be of interest to examine the impact of the Artemis-P70 mutation on lymphoid neoplasia alone and in the context of p53 mutation.
Summary
Together, our current findings provide insights into the molecular mechanisms that underlie partial combined immunodeficiency in patients caused by a hypomorphic Artemis allele. We provide evidence that the mutant Artemis-P70 nuclease is defective for DNA end processing during endogenous V(D)J recombination and exhibits reduced endonucleolytic activities despite retaining stable interaction with DNA-PKcs. Loss of the C terminus abrogates the ability of the Artemis-P70 protein to be phosphorylated by DNA-PKcs. Thus, although dispensable for activating endonucleolytic activities, our findings indicate that phosphorylation of the Artemis C terminus may play an important role in modulating in vivo functions. Importantly, we find that the Artemis-P70 mutation not only impairs end processing during V(D)J recombination but also leads to general DNA DSB repair defects that result in aberrant chromosomal rearrangements and genomic instability. Thus, partial loss of Artemis function may predispose to lymphoid or other malignancies in addition to causing combined immunodeficiencies.
MATERIALS AND METHODS
Generation of Artemis-P70 knock-in mice.
The targeting construct was generated using the pLNTK plasmid (pSalI–loxP-Neor-loxP–XhoI-TK cassette) (43). The 2.5-kbp 5′ homology arm and a 2.7-kbp 3′ homology arm were PCR amplified from R1 mouse ES cell genomic DNA. Both homology arms were subcloned into pLNTK and fully sequenced. The nucleotide changes to introduce the nonsense mutation at aa D449 in the mouse Artemis gene were introduced via site-directed mutagenesis (Agilent Technologies) and confirmed by sequence analysis. The targeting construct was linearized with PvuI and electroporated into 129Sv-derived R1 mouse ES cells. Individual ES clones were selected in G418 and counterselected in gancyclovir. Homologously targeted recombinants were identified by Southern blotting using EcoRV-digested genomic DNA and 5′ and 3′ genomic probes.
The successfully targeted ES cell lines were subcloned and subsequently karyotyped. Two independently derived subclones with normal karyotypes were injected into C57BL/6 blastocysts. The resulting chimeras were bred to C57BL/6 mice to obtain progeny with germline transmission of the Artemis-P70 mutation. The germline Art+/P70-Neo mice were subsequently bred to transgenic mice expressing the Cre recombinase from the EIIA promoter to delete the LoxP-flanked Neor gene during early embryogenesis. Progeny harboring the Neo-deleted mutant Artemis allele were screened by Southern blot analyses and then intercrossed to obtain Art+/+, Art+/P70, and ArtP70/P70 mice. Littermates were used for all experiments. The mice were housed in a room dedicated to immunodeficient mice within a specific pathogen-free facility. All mouse protocols were approved of by the University Committee on Use and Care of Animals at the University of Michigan, an Association for Assesment and Accreditation of Laboratory Animal Care, International accredited institute.
PCR analysis of IgH and TCR rearrangements.
DJH and DJβ rearrangements in sorted pro- and pre–B cells and purified DN thymocytes, respectively, were analyzed by PCR amplification as previously described (details in supplemental text) (44, 45). PCR analyses were repeated three times on at least two independent samples of genomic DNA. Extrachromosomal Dδ2-Jδ1 signal joints were analyzed as previously described (46). PCR analyses were repeated three times with two or three independent sets of genomic thymocyte DNA samples.
LM-PCR amplification of signal and coding ends.
Genomic DNA isolated from sorted Art+/+, ArtP70/P70, and Art−/− DN thymocytes was treated with T4 DNA polymerase, MBN, or left untreated and then ligated to double-stranded DNA linkers as previously described (details in supplemental text) (35). The ligation products were then subjected to nested PCR amplification reactions using locus-specific primers to detect either signal or coding V(D)J ends. The PCR products were electrophoresed through 6% polyacrylamide gels, transferred to zeta-probe membrane, and then probed with 32P 5′ end-labeled locus-specific oligonucleotides. Each experiment was repeated a minimum of three times with genomic DNA isolated from at least three different mice of each genotype.
Survival and genomic instability assays.
Passage 2 (P2) primary MEFs of the indicated genotypes were exposed to the indicated doses of γ rays from a 137Cs source and plated in duplicate at 2 × 105 cells per 6-cm gelatinized dish. The cells were cultured for 7 d, trypsinized, and stained with trypan blue. Live cells were counted using a hemacytometer. For bleomycin sensitivity, P2 MEFs were plated at 1.5 × 105 cells per 6-cm gelatinized dish and treated with the indicated doses of drug for 24 h. Cells were cultured for 4 d after the drug was removed, harvested, and live cells were counted. Percentage of survival was calculated as the number of treated cells over untreated control cells. Each experiment was repeated a minimum of three times with at least two independently derived cell lines.
P2 primary MEFs of the indicated genotypes were exposed to 200 rads of γ rays from a 137Cs source. After exposure, the cells were plated at 1.5 × 106 cells per 10-cm gelatinized dish and allowed to recover for 24 h. Cells were treated with colcemid for 4 h, harvested, and fixed. Metaphases were analyzed by DAPI staining. Data represent the results from two experiments performed on two independently derived cell lines.
Transient V(D)J recombination assay.
Transient V(D)J recombination assay was performed as previously described (details in supplemental text) (37).
In vitro phosphorylation assay.
Reaction mixtures contained 25 mM Hepes, pH 7.8, 50 mM KCl, 10 mM MgCl2, 2 mM EGTA, 1 mM DTT, 0.3 mg/ml sheared salmon sperm DNA, 0.14 µM γ-[32P]ATP, and purified WT or mutant Artemis proteins in a 30-µl volume (details in supplemental text). Kinase reactions were initiated upon addition of 350 ng of purified DNA-PKcs and were incubated at 37°C for 90 min. Reactions were stopped upon addition of SDS/EDTA and proteins were fractionated on a 10% SDS-PAGE gel. The gels were then dried and images were obtained by autoradiography.
DNA nuclease assays.
The single-strand DNA nuclease assay was performed in reaction mixtures containing 50–100 ng Artemis WT and mutant proteins, 0.25 pmol 32P 5′ end-labeled 20-nt oligo-dA substrate, 50 mM Tris, pH 8.0, 10 mM MgCl2, 20 mM NaCl, 0.25 mM ATP, 0.05 µg/µl BSA, and 1 mM DTT in a 50-µl volume. The mixtures were incubated at 37°C for 2 h, stopped with EDTA and formamide, and then loaded onto a 20% denaturing acrylamide gel. The hairpin endonuclease assays were performed in reaction mixture containing 25 mM Tris, pH 8.0, 10 mM MgCl2, or 10 mM MnCl2 (where indicated), 0.125 pmol 32P 5′ end-labeled hairpin DNA substrate, 0.05 µg/µl BSA, 0.25 mM ATP, and 0.25 µM of a 32-bp double-stranded DNA oligonucleotide in a 20-µl volume. Reactions were incubated at 37°C for 2 h. The 3′ overhang endonuclease assays were performed as described for hairpin nicking, except that 0.25 pmol 32P 5′ end-labeled 3′ overhang substrate was incubated with the purified Artemis proteins for 1 h. Where indicated, 350 ng DNA-PKcs was added to each reaction. Nuclease reactions were stopped with EDTA and formamide and then electrophoresed through a 17% denaturing acrylamide gel. Products were visualized by autoradiography. Each assay was repeated at least three times with two or more independent preparations of the purified Artemis proteins.
Online supplemental material.
Supplemental text contains a detailed description of the materials and methods and primer sequences. Fig. S1 shows sequences of endogenous IgH and TCR-β coding joints. Fig. S2 shows sequences of ApaLI-resistant RS joints. Fig. S3 shows sequences of coding joints recovered from transient transfection complementation V(D)J recombination assay in ES cells. Fig. S4 shows sequences of coding joints recovered from transient transfection V(D)J recombination assay in Artemis-P70 MEFs. Table S1 shows Mendelian inheritance of the Artemis-P70 allele. Table S2 shows frequency of ApaLI-sensitive Dδ2-Jδ1 signal joints. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082396/DC1.
Acknowledgments
This work was supported by National Institutes of Health grant AI063058 (NIAID), Pew Scholar's Award (Pew Charitable Trusts), Munn IDEA award (UM Cancer Center) to J. Sekiguchi, and in part by the National Institutes of Health UM Cancer Center Support Grant (5 P30 CA46592). M. Kubec is supported by the National Institutes of Health Genetics Training grant T32-GM07544 (NIGMS).
The authors have no conflicting interests.
Footnotes
Abbreviations used: cDNA, complementary DNA; DN, double negative; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DP, double positive; DSB, double-strand break; ES, embryonic stem; IgH, Ig heavy chain; IR, ionizing radiation; LM-PCR, ligation-mediated PCR; MBN, mung bean nuclease; MEF, mouse embryonic fibroblast; mRNA, messenger RNA; NHEJ, nonhomologous end joining; RAG, recombination activating gene; RSS, recombination signal sequence; RS-SCID, radiosensitive T−B− severe combined immunodeficiency.
References
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination.Cell. 109:S45–S55 [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Alt F.W., Oettinger M. 2004. The mechanism of V(D)J recombination. Molecular Biology of B cells. Alt F.W., Honjo T., Academic Press, San Diego, CA: 57–78 [Google Scholar]
- Fugmann S.D. 2001. RAG1 and RAG2 in V(D)J recombination and transposition.Immunol. Res. 23:23–39 [DOI] [PubMed] [Google Scholar]
- Gellert M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation.Annu. Rev. Biochem. 71:101–132 [DOI] [PubMed] [Google Scholar]
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondaneche M.C., Sanal O., Plebani A., Stephan J.L., Hufnagel M., le Deist F., et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly.Cell. 124:287–299 [DOI] [PubMed] [Google Scholar]
- Ahnesorg P., Smith P., Jackson S.P. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining.Cell. 124:301–313 [DOI] [PubMed] [Google Scholar]
- Ma Y., Pannicke U., Schwarz K., Lieber M.R. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination.Cell. 108:781–794 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Yamagishi H. 1992. Biased reading frames of pre-existing DH–JH coding joints and preferential nucleotide insertions at VH–DJH signal joints of excision products of immunoglobulin heavy chain gene rearrangements.EMBO J. 11:4869–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias S., Muegge K., Durum S.K. 1996. Junctional diversity in signal joints from T cell receptor beta and delta loci via terminal deoxynucleotidyl transferase and exonucleolytic activity.J. Exp. Med. 184:1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T., Yamagishi H. 1992. Novel excision products of T cell receptor gamma gene rearrangements and developmental stage specificity implied by the frequency of nucleotide insertions at signal joints.Eur. J. Immunol. 22:101–106 [DOI] [PubMed] [Google Scholar]
- Touvrey C., Couedel C., Soulas P., Couderc R., Jasin M., de Villartay J.P., Marche P.N., Jouvin-Marche E., Candeias S.M. 2008. Distinct effects of DNA-PKcs and Artemis inactivation on signal joint formation in vivo.Mol. Immunol. 45:3383–3391 [DOI] [PubMed] [Google Scholar]
- Touvrey C., Borel E., Marche P.N., Jouvin-Marche E., Candeias S.M. 2006. Gene-specific signal joint modifications during V(D)J recombination of TCRAD locus genes in murine and human thymocytes.Immunobiology. 211:741–751 [DOI] [PubMed] [Google Scholar]
- Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency.Cell. 105:177–186 [DOI] [PubMed] [Google Scholar]
- Le Deist F., Poinsignon C., Moshous D., Fischer A., de Villartay J.P. 2004. Artemis sheds new light on V(D)J recombination.Immunol. Rev. 200:142–155 [DOI] [PubMed] [Google Scholar]
- Moshous D., Pannetier C., Chasseval Rd R., Deist Fl F., Cavazzana-Calvo M., Romana S., Macintyre E., Canioni D., Brousse N., Fischer A., et al. 2003. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis.J. Clin. Invest. 111:381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege M., Ma Y., Manfras B., Kalwak K., Lu H., Lieber M.R., Schwarz K., Pannicke U. 2005. Omenn syndrome due to ARTEMIS mutations.Blood. 105:4179–4186 [DOI] [PubMed] [Google Scholar]
- van der Burg M., Verkaik N.S., den Dekker A.T., Barendregt B.H., Pico-Knijnenburg I., Tezcan I., vanDongen J.J., van Gent D.C. 2007. Defective Artemis nuclease is characterized by coding joints with microhomology in long palindromic-nucleotide stretches.Eur. J. Immunol. 37:3522–3528 [DOI] [PubMed] [Google Scholar]
- Musio A., Marrella V., Sobacchi C., Rucci F., Fariselli L., Giliani S., Lanzi G., Notarangelo L.D., Delia D., Colombo R., et al. 2005. Damaging-agent sensitivity of Artemis-deficient cell lines.Eur. J. Immunol. 35:1250–1256 [DOI] [PubMed] [Google Scholar]
- Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M.R. 2005. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis.J. Biol. Chem. 280:33839–33846 [DOI] [PubMed] [Google Scholar]
- Niewolik D., Pannicke U., Lu H., Ma Y., Wang L.C., Kulesza P., Zandi E., Lieber M.R., Schwarz K. 2006. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs.J. Biol. Chem. 281:33900–33909 [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., Yu Y., Riballo E., Douglas P., Walker S.A., Ye R., Harer C., Marchetti C., Morrice N., Jeggo P.A., Lees-Miller S.P. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity.EMBO J. 25:3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand S., Pope L., De Chasseval R., Gosselin D., Dong F., de Villartay J.P., Hache R.J. 2006. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin.J. Mol. Biol. 358:1200–1211 [DOI] [PubMed] [Google Scholar]
- Poinsignon C., Moshous D., Callebaut I., de Chasseval R., Villey I., de Villartay J.P. 2004. The metallo-β-lactamase/β-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination.J. Exp. Med. 199:315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S., Sekiguchi J., Zhu C., Cheng H.L., Manis J., Whitlow S., DeVido J., Foy D., Chaudhuri J., Lombard D., Alt F.W. 2002. Leaky scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice.Mol. Cell. 10:1379–1390 [DOI] [PubMed] [Google Scholar]
- Li G., Alt F.W., Cheng H.L., Brush J.W., Goff P.H., Murphy M.M., Franco S., Zhang Y., Zha S. 2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination.Mol. Cell. 31:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D. 1993. Mice lacking TdT: mature animals with an immature lymphocyte repertoire.Science. 261:1175–1178 [DOI] [PubMed] [Google Scholar]
- Komori T., Okada A., Stewart V., Alt F.W. 1993. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes.Science. 261:1171–1175 [DOI] [PubMed] [Google Scholar]
- Liston A., Enders A., Siggs O.M. 2008. Unravelling the association of partial T-cell immunodeficiency and immune dysregulation.Nat. Rev. Immunol. 8:545–558 [DOI] [PubMed] [Google Scholar]
- Gras S., Kjer-Nielsen L., Burrows S.R., McCluskey J., Rossjohn J. 2008. T-cell receptor bias and immunity.Curr. Opin. Immunol. 20:119–125 [DOI] [PubMed] [Google Scholar]
- Turner S.J., Doherty P.C., McCluskey J., Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity.Nat. Rev. Immunol. 6:883–894 [DOI] [PubMed] [Google Scholar]
- Akira S., Okazaki K., Sakano H. 1987. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining.Science. 238:1134–1138 [DOI] [PubMed] [Google Scholar]
- Hesse J.E., Lieber M.R., Mizuuchi K., Gellert M. 1989. V(D)J recombination: a functional definition of the joining signals.Genes Dev. 3:1053–1061 [DOI] [PubMed] [Google Scholar]
- Roth D.B., Menetski J.P., Nakajima P.B., Bosma M.J., Gellert M. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes.Cell. 70:983–991 [DOI] [PubMed] [Google Scholar]
- Zhu C., Roth D.B. 1995. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination.Immunity. 2:101–112 [DOI] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu A., Morrow T., Baxter M., Peng A. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated.Genes Dev. 7:2520–2532 [DOI] [PubMed] [Google Scholar]
- Roth D.B., Zhu C., Gellert M. 1993. Characterization of broken DNA molecules associated with V(D)J recombination.Proc. Natl. Acad. Sci. USA. 90:10788–10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse J.E., Lieber M.R., Gellert M., Mizuuchi K. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals.Cell. 49:775–783 [DOI] [PubMed] [Google Scholar]
- Rooney S., Alt F.W., Lombard D., Whitlow S., Eckersdorff M., Fleming J., Fugmann S., Ferguson D.O., Schatz D.G., Sekiguchi J. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells.J. Exp. Med. 197:553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet J., Frit P., Delteil C., de Villartay J.P., Salles B., Calsou P. 2006. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends.J. Biol. Chem. 281:27784–27793 [DOI] [PubMed] [Google Scholar]
- Nickoloff J.A., De Haro L.P., Wray J., Hromas R. 2008. Mechanisms of leukemia translocations.Curr. Opin. Hematol. 15:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S., Sekiguchi J., Whitlow S., Eckersdorff M., Manis J.P., Lee C., Ferguson D.O., Alt F.W. 2004. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells.Proc. Natl. Acad. Sci. USA. 101:2410–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.D., Ferguson D.O., Alt F.W. 2003. The role of DNA breaks in genomic instability and tumorigenesis.Immunol. Rev. 194:77–95 [DOI] [PubMed] [Google Scholar]
- Gorman J.R., van der Stoep N., Monroe R., Cogne M., Davidson L., Alt F.W. 1996. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes.Immunity. 5:241–252 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., Baltimore D. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription.J. Exp. Med. 173:711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner F., Alt F.W., Monroe R., Chu M., Sleckman B.P., Davidson L., Swat W. 1999. Immature thymocytes employ distinct signaling pathways for allelic exclusion versus differentiation and expansion.Immunity. 10:537–546 [DOI] [PubMed] [Google Scholar]
- Zhu C., Bogue M.A., Lim D.S., Hasty P., Roth D.B. 1996. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates.Cell. 86:379–389 [DOI] [PubMed] [Google Scholar]