Abstract
Revertants of a colcemid resistant Chinese hamster ovary cell line with an altered (D45Y) β-tubulin have allowed the identification of 4 cis-acting mutations (L187R, Y398C, a 12 amino acid in-frame deletion, and a C-terminal truncation) that act by destabilizing the mutant tubulin and preventing it from incorporating into microtubules. These unstable β-tubulins fail to form heterodimers and are predominantly found in association with the chaperonin CCT, suggesting that they cannot undergo productive folding. In agreement with these in vivo observations, we show that the defective β-tubulins do not stably interact with cofactors involved in the tubulin folding pathway and hence fail to exchange with β-tubulin in purified αβ heterodimers. Treatment of cells with MG132 causes an accumulation of the aberrant tubulins, indicating that improperly folded β-tubulin is degraded by the proteasome. Rapid degradation of the mutant tubulin does not elicit compensatory changes in wild-type tubulin synthesis or assembly. Instead, loss of β-tubulin from the mutant allele causes a 30–40% decrease in cellular tubulin content with no obvious effect on cell growth or survival.
Microtubules are essential cytoskeletal structures responsible for much of the vesicular traffic in cells, organization of the endoplasmic reticulum and Golgi apparatus, and segregation of sister chromatids prior to cell division. They assemble by the endwise concatenation of αβ tubulin heterodimers to form protofilaments, 13 of which associate laterally to form the walls of microtubules in most eukaryotic cells. Alpha and beta tubulin subunits each follow a complex folding pathway before joining to form assembly competent heterodimers. This process involves prefoldin (1,2), CCT1 (cytosolic chaperonin containing TCP-1, also called TCP-1 complexes, TriC, or cytosolic chaperonin) (3–5), and the downstream participation of a series of protein cofactors (6–9). Prefoldin binds to nascent chains and is thought to target newly synthesized tubulin to CCT (1) which then releases these polypeptides in a quasi-native state. The released β-tubulin is captured by either cofactor A or cofactor D. In a parallel pathway, quasi-native α-tubulin polypeptides are captured by either cofactor B or cofactor E. Cofactor E/α and cofactor D/β complexes interact to form a supercomplex. Entry of cofactor C into this supercomplex triggers GTP hydrolysis by β-tubulin, a reaction that results in the release of newly formed assembly competent αβ tubulin heterodimers (6,7).
Mammalian cells express multiple tubulin genes, each of which encodes a distinct α- or β-tubulin isotype (10,11). As far as is known, these distinct gene products follow a common folding pathway. In addition to their role in the de novo assembly of tubulin dimers, cofactors C, D and E act in concert on native heterodimers as GAPs. This reaction occurs at a tubulin concentration far below that required for polymerization into microtubules, and is thought to act primarily as a quality control mechanism, constantly checking the capacity of heterodimers to productively hydrolyze GTP (12). This mechanism would protect the cell from the potentially deleterious consequences of incorporating GTPase-defective subunits into microtubules; however, the means whereby defectively folded tubulin polypeptides are recognized and targeted for degradation is unknown.
A new strategy for examining tubulin degradation has emerged from genetic studies. We and others have described a growing number of tubulin mutations that confer resistance to antimitotic drugs in mammalian cells and that act by altering microtubule assembly in a manner that counteracts the action of the drugs (13,14). One of these resistant cell lines, Cmd 4, has a D45Y mutation in one allele of its class I β-tubulin (hereafter called β1-tubulin) gene that produces hyperstable microtubules, colcemid resistance, hypersensitivity to paclitaxel, and temperature sensitivity for growth (15–17). In recent studies, we analyzed revertants of Cmd 4 and reported that reversion of colcemid resistance can occur by cis-acting suppressor mutations that destabilize microtubules, by mutations that increase the affinity of tubulin for colcemid, or by mutations that prevent expression of the mutant allele (18,19). Here we report that some revertants produce functionally inactive mutant β1-tubulin because of intra-allelic mutations that interfere with the folding of the protein. We show that misfolded tubulin fails to interact with cofactors A and D following its release from CCT, and is rapidly degraded by the proteasome resulting in a large decrease in cellular tubulin content but no effect on growth.
EXPERIMENTAL PROCEDURES
Constructs and Reagents
pRCHAβ1 is a pRC/CMV plasmid (Invitrogen, Carlsbad, CA) containing a neomycin resistance gene driven by a constitutive SV40 promoter, and a β1 tubulin cDNA (GenBank Accession no. U08342) fused at the 3’ end to a sequence encoding a 9 amino acid hemagglutinin (HA) epitope under the control of a CMV promoter. The plasmid also contains a T7 promoter that allows it to drive in vitro transcription of cloned sequences (see below). For some experiments, mutation of the HAβ1-tubulin cDNA was carried out using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). pGFP-CoD contains tubulin folding cofactor D cDNA (GenBank Accession no. U61233) fused to the 3’ end of GFP. MG132 was purchased from Peptides International, Inc, Louisville, KY. Rat anti-TCP-1α (CCTα) monoclonal antibody was from StressGen Biotechnologies, Inc., Victoria, BC. Bovine brain tubulin was from Cytoskeleton Inc., Boulder, CO. TnT quick coupled transcription/translation system was from Promega (Madison, WI). Antibodies recognizing the amino terminus of β-tubulin, the C-terminus of tyrosinated α-tubulin, and GFP were generous gifts from Dr. Don Cleveland, Dr. Jeanette Bulinski, and Dr. William Margolin respectively.
Growth and Derivation of Cell Lines
Cells were grown in alpha modification of minimum essential medium (Sigma-Aldrich, St. Louis, MO) containing 50 U/ml penicillin, 50 µg/ml streptomycin, and 5% fetal bovine serum (Atlanta Biologicals, Atlanta, GA) at 37 °C and 5% CO2. Cmd 4 is a colcemid resistant and temperature sensitive Chinese hamster ovary (CHO) cell mutant that was previously described (15,16). Spontaneous revertants 6H2, 6H3, A5, and 5L1 were isolated from Cmd 4 by selecting cells able to grow at the non-permissive temperature of 40.5 °C (20).
Two-Dimensional Gel Electrophoresis
Cells were labeled for 30 min with 20 µCi/ml Tran [35S] label (mixture of methionine and cysteine, 1000 Ci/mmol; ICN Biomedicals, Costa Mesa, CA) in methionine-free minimum essential medium (Sigma-Aldrich) and lysed with hot (100 °C) SDS dissociation buffer (21). Solubilized proteins were precipitated with 5 volumes of cold acetone, and the protein pellets were resolubilized in urea sample buffer and analyzed by 2D gels as previously described (22).
Transfection, Immunofluorescence and Proteasomal Inhibition Experiments
CHO 10001 cells (16) were transfected with wild-type or mutant pRCHAβ1 using LipofectAMINE (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The efficiency of cell-to-cell expression was monitored 24 h post-transfection using immunofluorescence. The cells were rinsed with PBS, fixed with methanol at −20 °C, rehydrated with PBS, and incubated with a 1: 100 dilution of a rabbit antibody specific for the HA tag (Bethyl Laboratories, Montgomery, TX) followed by a 1:50 dilution of Alexa 488-conjugated goat anti-rabbit IgG (Invitrogen).
To inhibit proteasomal degradation of mutant tubulin, stably transfected cells were isolated in 2 mg/ml G418 (geneticin, Cellgro Inc., Herndon, VA) and incubated with or without 25 µM MG132 for 8 hours at 37 °C. Cells were then lysed in hot (100 °C) SDS, and proteins were separated by SDS-PAGE and transferred onto nitrocellulose (23). The transfected mutant tubulin was probed with anti-HA tag monoclonal antibody 12CA5 (Roche, Indianapolis, IN). Immunoreactive bands were detected using a 1:2000 dilution of peroxidase-conjugated goat anti-mouse IgG (Sigma-Aldrich) and SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, IL).
Measurement of Assembled Tubulin
To measure the distribution of tubulin between nonassembled and polymerized pools, cells were grown to near confluence in 24-well dishes and lysed with microtubule buffer (MTB) (20 mM Tris–HCl, pH 6.8, 0.5% Nonidet P- 40, 1 mM MgCl2, 2 mM EGTA) containing 0.14 M NaCl and 4 µg/ml paclitaxel. Cell remnants scraped from the dish in this buffer were transferred to microcentrifuge tubes and fractionated into soluble and sedimentable proteins by centrifugation at 12,000 g and 4 °C as previously described (24). Tubulin was quantified by first adding an equal volume of an E. coli SDS cell lysate containing a GST-α-tubulin fusion protein to each fraction, precipitating the proteins with acetone, and redissolving the precipitated proteins with SDS sample buffer (18). Proteins were then separated by 7.5% SDS-PAGE and transferred onto nitrocellulose (Schleicher and Schuell, Keene, NH). The tubulin in each fraction was detected using a monoclonal antibody to α-tubulin (1:2000 dilution of DM1A, Sigma-Aldrich) followed by cy5-conjugated goat antimouse IgG (1:2000 dilution, Chemicon International Inc., Temecula, CA). Fluorescence images were acquired using a Storm image analyzer (Molecular Dynamics) and bands were quantified using NIH Image (http://rsb.info.nih.gov/nih-image). Alpha tubulin was normalized by dividing by the GST-α-tubulin in the same fraction, and the percent of tubulin that was polymerized was calculated using the equation %P = [(α/GSTα)P/(α/GSTα)P+S] X 100%, where P denotes the α-tubulin to GST-α-tubulin ratio in the pellet and P+S denotes the sum of the ratios for pellet and supernatant fractions.
DNA Sequencing
Revertant cells were grown to 80% confluence in 60 mm dishes, rinsed three times with phosphate buffered saline, lysed with K buffer (20 mM Tris–HCl, pH 8.0, 50 mM KCl, 2.5 mM MgCl2, 0.5% Tween 20), and digested at 55 °C with 2 mg/ml proteinase K (Sigma-Aldrich) for 45 min. The proteinase K was then inactivated by heating to 100 °C for 10 min. This genomic DNA was utilized as a template to amplify different regions of the β1-tubulin gene using pairs of primers homologous to translated and untranslated sequences of the DNA. The PCR amplified DNA was directly sequenced (DNA Sequencing Core Facility, Baylor College of Medicine, Houston, TX).
Measurement of Tubulin at Steady State
To measure α-tubulin levels, cells were grown to near confluence in 24 well dishes and lysed with 1% SDS. Solubilized proteins were precipitated with 5 volumes of acetone, DNA was removed, and the samples were centrifuged for 5 min at 12,000 g. Proteins in the pellet were resolubilized in SDS sample buffer, separated by 7.5% SDS-PAGE, and transferred to nitrocellulose. α- Tubulin and actin were detected by incubating the blot with mouse monoclonal antibodies DM1A (1:1000 dilution, Sigma-Aldrich) and C4 (1:50,000 dilution, Chemicon) followed by a Cy5-conjugated goat antimouse IgG (1:2000 dilution, Chemicon). Fluorescent images were acquired with a Storm Imager as described above. The α-tubulin in each sample was normalized by dividing by the actin signal, and compared to wild type cells set at 100%. Each experiment was repeated at least 3 times.
Glycerol Gradient Sedimentation
Near confluent 35 mm dishes of mutant cell lines 6H2 and 6H3 were lysed in 200 µl MTB containing 0.14 M NaCl and a protease inhibitor cocktail (Sigma-Aldrich), and then centrifuged at 12,000 g for 15 min. The supernatants were loaded onto 10 ml 5–40% glycerol gradients and centrifuged in a swinging bucket rotor at 100,000 g for 16 hours at 4 °C. Fractions were resolved by SDS gel electrophoresis, transferred to nitrocellulose, and probed with either a mouse anti-β-tubulin N-terminal antibody or a rat anti-TCP-1α antibody.
Immunoprecipitation
To search for interactions of mutant β-tubulin with α-tubulin, cell lines 6H2 and 6H3, which produce mutant β-tubulins with a decreased apparent molecular mass, were lysed in MTB containing 0.14 M NaCl and centrifuged at 12,000 g for 15 min at 4 °C. The clarified supernatants were incubated with protein G Sepharose (Amersham) prebound with rabbit antibody to tyrosinated α-tubulin, and the beads were centrifuged, washed with the same MTB buffer, and stripped with SDS sample buffer. The immunoprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with a mouse monoclonal antibody to the N-terminus of β-tubulin.
To detect potential interactions between β-tubulin and cofactor D, 6H3 was transfected with pGFP-CoD, and wild type cells were cotransfected with pGFP-CoD and pRC/CMV containing wild-type or mutant (6H2, Y398C) HAβ1-tubulin cDNA. At 36 h post-transfection, cell extracts were prepared as described above and incubated with protein G Sepharose prebound with rabbit antibody to GFP. The immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose. For 6H3 the western blot was probed with a mouse monoclonal antibody to the N-terminus of β-tubulin; in the case of other cell lines, mouse monoclonal antibody 12CA5 (Roche) to the HA tag was used to distinguish transfected tubulin from the co-migrating endogenous tubulin.
In Vitro Transcription and Translation
Equal amounts of β1-tubulin cDNAs with or without mutations were transcribed and translated in vitro for 90 min in the presence of [35S]methionine using a TnT kit (Promega). The radiolabeled in vitro synthesized proteins were further incubated for 30 min at 37 °C, in the presence or absence of purified bovine brain tubulin (Cytoskeleton, Inc., Boulder, CO [0.25 mg/ml, final concentration]), and analyzed by electrophoresis under non-denaturing conditions (4). A portion of each sample was also resolved by SDS-PAGE to demonstrate that all tubulins were efficiently synthesized in the TnT system.
Reconstituted In Vitro Folding
Wild-type and mutant HAβ1-tubulin cDNAs were cloned into a pET28a+ bacterial expression vector (EMD Biosciences, Inc., Madison, WI), and [35S]methionine labeled proteins were produced in E. coli BL21DE3 cells (25). Inclusion bodies containing the radiolabeled tubulin were purified and solubilized in 8 M urea as described previously (4,6). These labeled target proteins were folded in vitro in reactions that contained purified CCT and one or more of the components required for reconstitution of the tubulin folding pathway (7). Reaction products were resolved on 4.5% native polyacrylamide gels (4).
RESULTS
Mutations Affecting the Assembly and Stability of β-Tubulin
Vertebrates express 7 β-tubulin genes that encode closely related, but distinct tubulin proteins or isotypes (10). Based on their C-terminal sequences, these isotypes have been categorized into classes (I, II, III, IVa, IVb, V, and VI), some of which are ubiquitously expressed while others are tissue specific (11,26). CHO cells express 3 of these isotypes (I, IVb, and V) in a ratio of 70:25:5 (27,28). Cmd 4, a colcemid resistant CHO cell line with increased sensitivity to paclitaxel and temperature sensitivity for growth (15,16), has a D45Y mutation in one copy of its class I β-tubulin gene such that 35% of its total β-tubulin is altered. Incorporation of this altered tubulin increases microtubule stability leading to a higher fraction of cellular tubulin in the polymer pool and an increase in microtubule bundle formation (17). Spontaneous revertants of Cmd 4 able to grow at the nonpermissive temperature coreverted to colcemid sensitivity at high frequency, showing that the two phenotypes are linked (20).
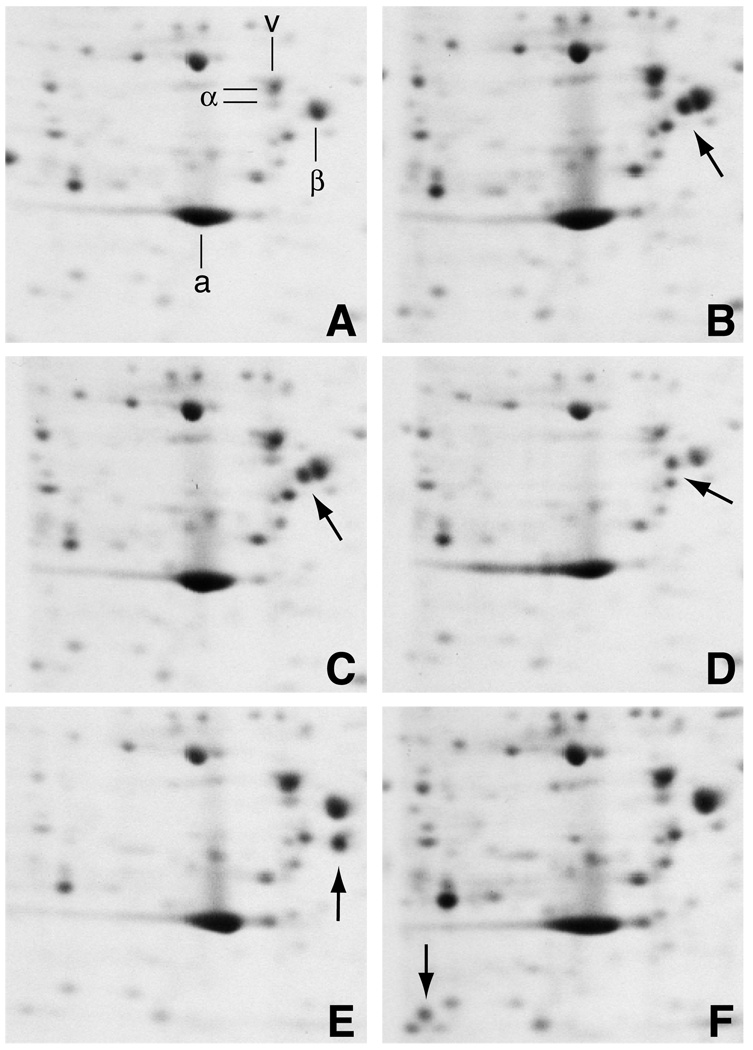
Two-dimensional gel analysis of extracts from four revertant cell lines that were pulse labeled for 30 min with [35S]methionine demonstrated that a mutant β-tubulin species was made in these cells and in Cmd 4 (Fig. 1). Whereas wild-type cells exhibited a single spot comprising all of the expressed β-tubulin isotypes (Fig. 1A), Cmd 4 produced a second, more basic β-tubulin consistent with its D45Y mutation (arrow, Fig. 1B). This mutant tubulin had undergone further alterations in revertants A5 (arrow, Fig. 1D), 6H2 (arrow, Fig. 1E), and 6H3 (arrow, Fig. 1F) as evidenced by the fact that the protein occupied a new position on the gel in each case. The identities of these new spots in the gel as mutant forms of β-tubulin were confirmed by peptide mapping (20). Revertant 5L1 (arrow, Fig. 1C) produced a mutant tubulin that migrated with the same mobility as the mutant tubulin in Cmd 4. Nevertheless, we suspected a second alteration in this tubulin because, like the other revertants, the protein spot was missing when the gels were stained for accumulated protein or radiolabeled to steady state (data not shown). Thus, unlike Cmd 4 which produces a stable mutant protein, the mutant β-tubulin in each of the revertant cell lines is synthesized at a normal rate, but is unstable and fails to accumulate to appreciable steady state levels. Pulse-chase experiments with strain 6H2 indicated that wild-type β-tubulin has a half life of over 50 h, consistent with previous estimates, but the mutant β1-tubulin has a half life of only 1–2 h (20,29).
Fig. 1.
Two-dimensional gel electrophoresis of mutant and revertant cell lines. Wild-type (A), Cmd 4 (B), 5L1 (C), A5 (D), 6H2 (E), and 6H3 (F) cell lines were metabolically labeled for 30 min with [35S]methionine, lysed in SDS, and resolved on 2D gels. Portions of the autoradiograms containing the major cytoskeletal proteins actin (a), vimentin (v), α-tubulin (α), and β-tubulin (β) are shown. Arrows indicate the presence of mutant β-tubulins. Gels are oriented with the basic end to the left.
To confirm the presence of additional mutations, genomic DNA was sequenced; the results are summarized in Table I. Consistent with the behavior of β-tubulin on 2D gels, revertants were found to have the original D45Y mutation but also possessed an electrophoretically silent Y398C mutation (5L1), a basic L187R mutation (A5), an in-frame deletion of amino acids T351-K362 (6H2), or a C-terminal truncation of approximately 120 amino acids (6H3 [see (30)]).
Table I.
Properties of Cmd 4 and its revertants
| Cell line | Cmd sensitivitya | Mutation | 2D gel shiftb |
|---|---|---|---|
| Wild-type | S | None | β |
| Cmd 4 | R | D45Y | ββ* |
| A5 | S | D45Y/L187R | ββ** |
| 5L1 | S | D45Y/Y398C | ββ* |
| 6H2 | S | D45Y/Δ351–362 | ββΔ |
| c6H3 | S | D45Y/Δ325–444 | ββΔ |
S, normal sensitivity to the drug; R, resistant.
Asterisk denotes a new spot with a 1 charge basic shift in migration; double asterisk denotes a 2 charge basic shift; Δ denotes a deletion causing a lower apparent molecular mass.
The size of the deletion is approximate.
Failure to Assemble Into Microtubules
We previously used a biochemical fractionation assay to demonstrate that newly synthesized mutant β-tubulin in revertant cell lines does not significantly sediment with microtubules (20). To extend these observations to an in vivo context, we visually monitored incorporation of the mutant tubulins following their transient overexpression in cultured cells. In addition, we eliminated possible contributions from the original D45Y substitution to the assembly defect (see Table I), by introducing the point mutations Y398C and L187R, and the 12 amino acid deletion characteristic of revertants 5L1, A5, and 6H2 into a wild-type β1-tubulin cDNA using site-directed mutagenesis. The β1-tubulin cDNA also contained a 3’ sequence encoding a C-terminal hemagglutinin (HA) tag so that the transfected tubulin could be easily distinguished from its endogenous wild-type counterpart.
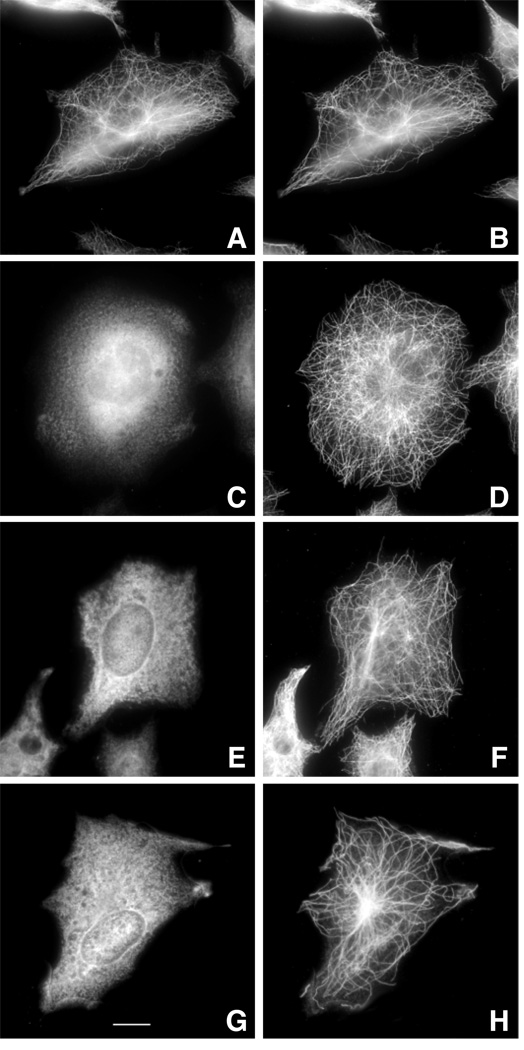
Immunofluorescence of cells transiently transfected with each of the cDNAs is shown in Fig. 2. Wild-type HAβ1-tubulin efficiently assembled into the microtubule cytoskeleton (Fig. 2A) and had a distribution identical to the endogenous tubulin (Fig. 2B). In contrast, all 3 of the mutant HAβ1-tubulins produced diffuse fluorescence with no evidence of incorporation into microtubules. We conclude that the secondary mutations alone are sufficient to preclude assembly of β-tubulin.
Fig. 2.
Immunofluorescence of transfected cells. Wild-type CHO cells were transfected with wild-type HAβ1-tubulin cDNA (A, B) or with the same DNA containing the 12 amino acid deletion from revertant 6H2 (C, D), the L187R mutation from revertant A5 (E, F), or the Y398C mutation from revertant 5L1 (G, H). At 24 h post-transfection, the cells were fixed with methanol and stained with antibodies to the HA tag (A, C, E, G) and α-tubulin (B, D, F, H). Assembly defective mutant 6H3 was not analyzed in this figure and some of the subsequent figures because we do not know the exact sequence of the truncated gene and therefore cannot construct the appropriate plasmid. The bar in panel G represents 10 µm.
Steady State Tubulin Levels are Reduced in Revertant Cell Lines
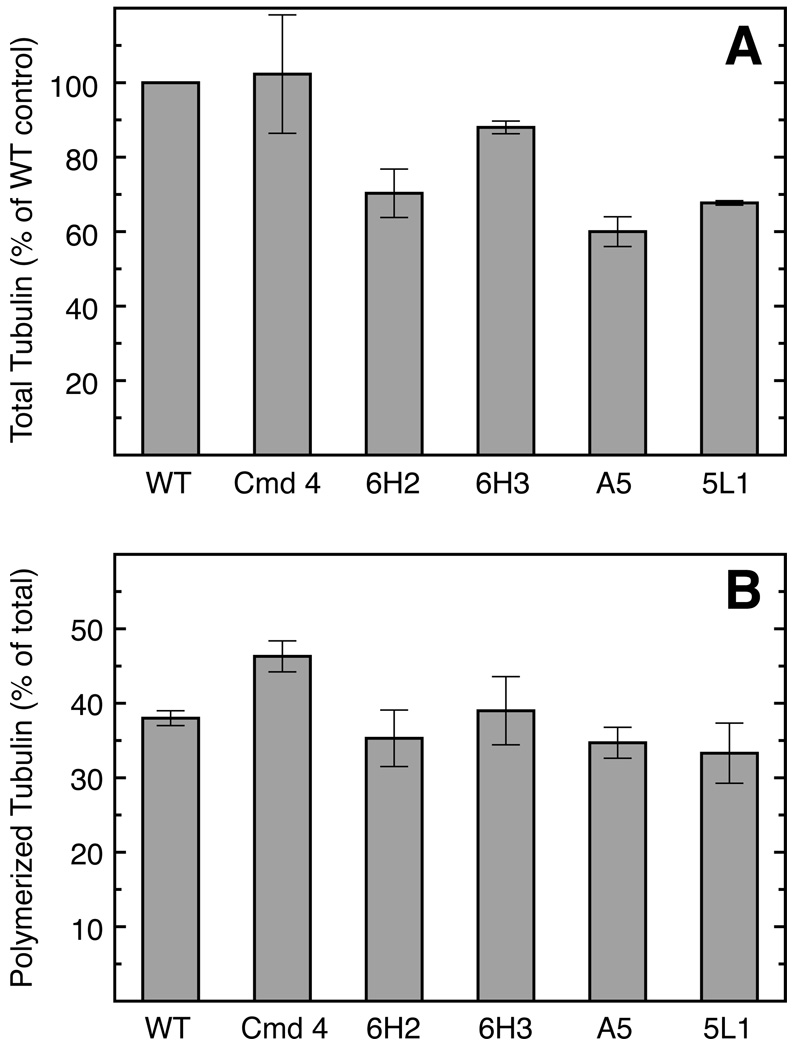
The presence of assembly defective and unstable β-tubulin accounting for 35% of the total could produce at least 2 potential outcomes: a reduced steady state amount of tubulin in revertant cell lines, or compensation for the loss of assembly defective tubulin by increased expression of wild-type tubulin genes. Using a radiolabeling approach, we previously showed that α- and β-tubulin levels are diminished in revertant 6H2 commeasurate with the loss of the assembly defective β1-tubulin (20). We confirmed these 6H2 results using an immunological approach to measure the relative α-tubulin content of the cells, and found that all the revertant cell lines with unstable assembly defective β1-tubulin display a similar 30–40% decrease in tubulin levels (Fig. 3A). The magnitude of this decrease is consistent with the expected 35% contribution of the mutant allele to β-tubulin production. We conclude that revertant CHO cells do not compensate for the loss of assembly defective β1-tubulin by up-regulating the expression of other β-tubulin genes and that α-tubulin is correspondingly reduced to a level equivalent to that of the surviving β-tubulin.
Fig. 3.
A, Relative tubulin levels. SDS lysates of the indicated cell lines were analyzed by Western blotting using anti-α-tubulin and anti-actin mouse monoclonal antibodies followed by a Cy5-conjugated goat anti-mouse IgG. Immunoreactive bands were quantified by their fluorescence emission. The ratio of α-tubulin to actin was calculated for each cell line, divided by the ratio calculated for wild-type cells, and multiplied by 100. B, Tubulin polymerization. The extent of tubulin polymerization in each of the indicated cell lines was determined by separating polymerized from soluble tubulin by centrifugation (see “Experimental Procedures”), and quantifying tubulin in pellet and supernatant fractions by Western blot analysis. The fraction of total cellular tubulin recovered in the cytoskeletal pellet is shown for each cell line. Values represent the average of 3 experiments with the standard deviation of the mean shown by error bars.
Endogenous Tubulin Assembly is Unaffected by the Presence of Assembly Defective Mutant Proteins
Reduced tubulin levels in revertant cell lines suggest that they might contain less microtubule polymer than wild-type cells. To explore this issue, we measured the extent of tubulin assembly in each of the cell lines. We found that Cmd 4 maintains a higher fraction of tubulin in the microtubule polymer compared to wild-type cells (Fig. 3B), consistent with an increased stability of microtubules that incorporate the mutant D45Y β-tubulin. Each of the revertant cell lines, however, has a normal (wild-type) extent of microtubule assembly. The results are consistent with the rapid degradation of the mutant β1-tubulin in revertant cell lines, but they indicate that the reduced cellular tubulin content does not trigger any mechanism that increases tubulin assembly in order to re-establish wild-type levels of microtubule polymer. Thus, revertant cells proliferate normally with a 35% reduction in both soluble and polymerized tubulin.
Assembly Defective Tubulin Fails to Form αβ Heterodimers
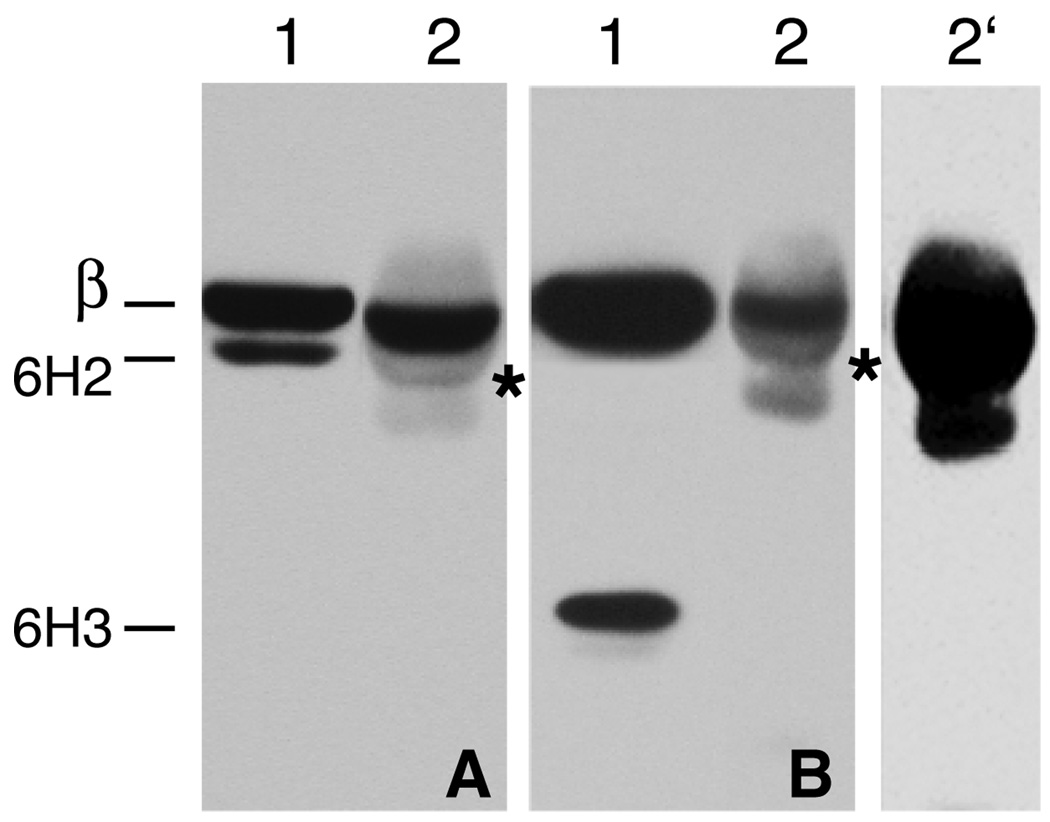
We considered two possibilities to account for the inability of mutant β1-tubulin to assemble in revertant cell lines. In one scenario, assembly defective β1-tubulin is synthesized but fails to form heterodimers with α–tubulin and is degraded as the free subunit. Alternatively, heterodimers may form, but they are recognized as assembly defective and degraded as dimers. To distinguish between these mechanisms, we immunoprecipitated α–tubulin and looked for coprecipitation of endogenous versus assembly defective β-tubulin. To simplify the analysis, the experiments were carried out with revertants 6H2 and 6H3 because these cell lines express assembly defective β1-tubulins that are easily detected because of their lower apparent molecular mass. Immunoprecipitation from 6H2 resulted in the pull down of endogenous wild-type, but not assembly defective β-tubulin (Fig. 4A, lane 2). Although a sharp weak band migrating slightly faster than the 6H2 assembly defective β1-tubulin was present in the immunoprecipitate (Fig. 4A, lane 2, asterisk), it is not likely to be related to the mutant protein because it was also evident in precipitations from 6H3 (Fig. 4B, lane 2). In addition, a broad diffuse band was seen comigrating with β-tubulin and is probably a result of weak cross-reactivity of the secondary goat anti-mouse IgG with rabbit IgG heavy chain.
Fig. 4.
Immunoprecipitation of tubulin heterodimers. Cell extracts from revertants 6H2 (panel A) and 6H3 (panel B) were incubated with a rabbit antibody to α-tubulin coupled to protein G Sepharose beads. The immunoprecipitates were analyzed by Western blotting with a mouse antibody specific for the N-terminus of β-tubulin. Lane 1 in each of the two panels represents the unfractionated cell extract and lane 2 represents the immunoprecipitate. Lane 2’ represents an overexposure of lane 2 from panel B to demonstrate that no 6H3 mutant tubulin is found in the immunoprecipitate. The light fuzzy bands and the weak sharp bands with a migration similar to 6H2 β-tubulin (asterisks) present in the immunoprecipitates are due to weak cross reactivity of the goat antimouse IgG secondary antibody with the rabbit IgG heavy chains. The migration of endogenous wild-type β-tubulin (β), 6H2 mutant β-tubulin (6H2) and mutant 6H3 β-tubulin (6H3) is indicated.
The results with 6H3 were more clear-cut because of the much larger molecular mass decrease in the assembly defective β1-tubulin (Fig. 4 B). Again, endogenous wild-type β-tubulin was co-immunoprecipitated with α-tubulin, but there was no trace of assembly defective β1-tubulin (lane 2) even when the gel was overexposed (lane 2’). We conclude that assembly defective β1-tubulin does not form heterodimers in vivo, at least for the 2 cell lines analyzed here.
Formation of a Complex With CCT
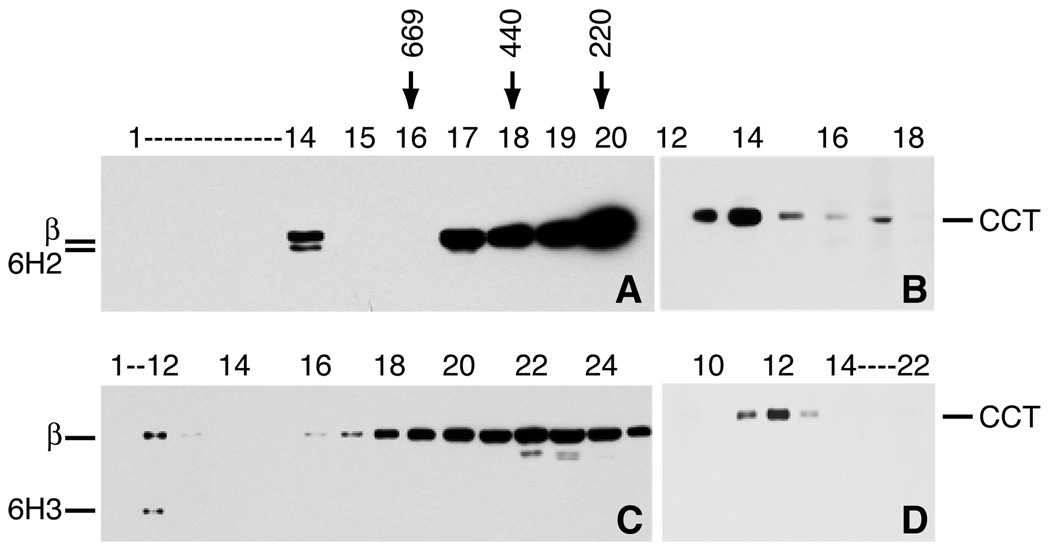
To determine whether assembly defective β1-tubulin exists in the cell as part of a larger complex, we analyzed its sedimentation behavior. Although the assembly defective protein did not sediment with the microtubule cytoskeleton at 12,000 g, a considerable portion of the protein did sediment at 100,000 g (data not shown). We therefore analyzed cell extracts from revertants 6H2 and 6H3 by glycerol gradient centrifugation. A western blot of fractions from the gradient using an antibody to the N-terminus of β-tubulin showed that wild-type β-tubulin in 6H2 (Fig. 5 A) and 6H3 (Fig. 5 C) stayed predominantly at the top of the gradient, but that assembly defective β1-tubulin migrated with a sedimentation velocity greater than that of thyroglobulin, a protein standard with a molecular mass of 669 kDa. The assembly defective tubulin appeared to be monodisperse and some of the wild-type β-tubulin also migrated in this fraction arguing that the increased sedimentation rate was not the result of aggregation, but rather due to its presence in a discreet complex. The size of the complex suggested the possibility that this fraction represented the interaction of both wild-type and assembly defective β1-tubulin with the chaperonin CCT which is obligatorily involved in the tubulin folding pathway (31,32). To test this possibility, the fractions were also tested with a monoclonal antibody to TCP-1α (Fig. 5 B, D). The same fraction that contained the assembly defective β1-tubulin stained heavily for TCP-1 α, suggesting that most of this tubulin is complexed with CCT. Assembly defective tubulin was not detected in any other fraction indicating that the protein might be targeted for degradation following its release from CCT.
Fig. 5.
Glycerol gradient centrifugation. Cellular extracts from revertants 6H2 (A, B) and 6H3 (C, D) were fractionated by glycerol gradient centrifugation. Fractions were analyzed by Western blotting with a mouse antibody to the N-terminus of β-tubulin (A, C) or a rat antibody to one of the subunits of the CCT complex (B, D). Arrows indicate the migration of thryroglobulin (669 kDa), ferritin (440 kDa), and catalase (232 kDa). Labeled bands include wild-type β-tubulin (β), 6H2 mutant β-tubulin (6H2), 6H3 mutant β-tubulin (6H3), and CCT subunit TCP-1α (CCT). Note that the fractions containing the mutant subunits (14 in panel A, 12 in panel C) are also the fractions that stain heavily for CCT.
Assembly Defective β1-tubulin Fails to Interact With Folding Cofactors In Vitro
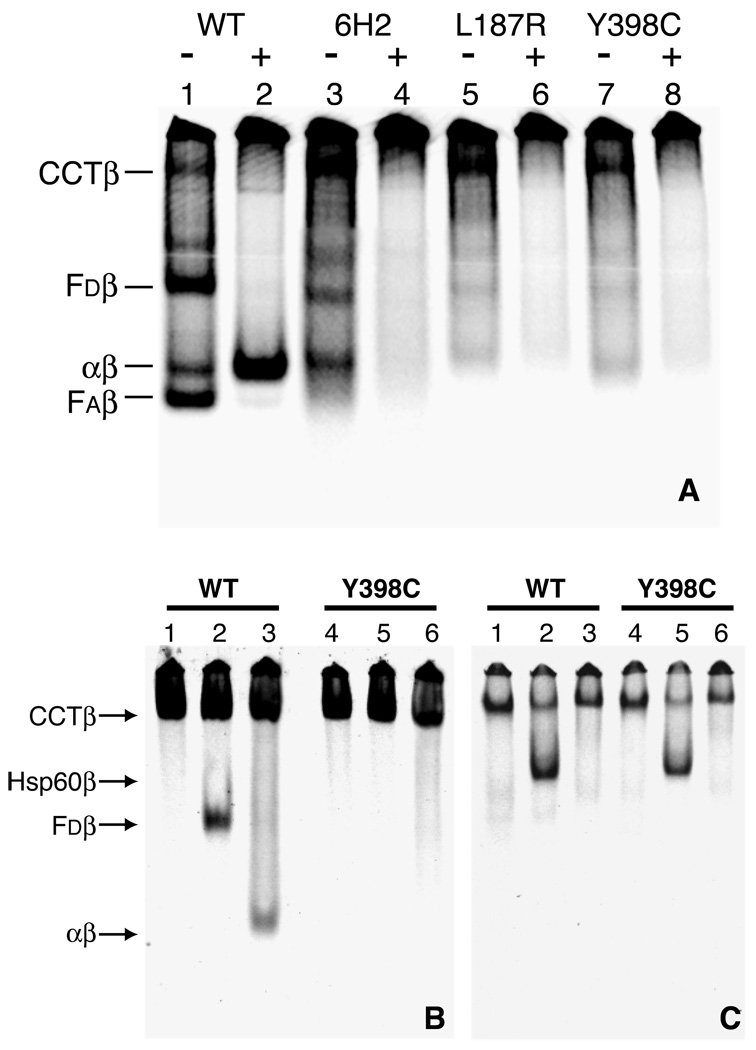
The in vivo fractionation data indicated that assembly defective β-tubulin is efficiently synthesized (Fig. 1), released from ribosomes, and targeted to CCT. Because its ratio relative to wild-type β-tubulin in the CCT complex mirrored the synthesis rates of the two proteins (compare Figure 1 and Figure 5), assembly defective β1-tubulin did not appear to be inappropriately retained in the CCT complex. We therefore looked for potential interactions of this tubulin with one or more of the cofactors that act downstream of CCT and that are involved in de novo heterodimer assembly. This was done both by translation in vitro, and in reconstituted folding assays using purified components. We found that cell-free synthesis of wild-type HAβ1-tubulin in a rabbit reticulocyte lysate (Fig. 6A, lane 1) generated bands indicative of complexes between β-tubulin and CCT, cofactor D, and cofactor A, as well as native α/β tubulin heterodimers (6). In contrast, assembly defective HAβ1-tubulins (Fig. 6A, lanes 3, 5, and 7) showed clear evidence for complex formation with CCT, but otherwise exhibited only relatively weak bands that did not correspond to the bands produced by HAβ1-tubulin.
Fig. 6.
In vitro synthesis and folding of β-tubulin. Panel A, A coupled transcription/translation system utilizing rabbit reticulocyte lysate was used to synthesize radiolabeled β-tubulin from plasmid DNA. Following synthesis, reaction mixtures were incubated with (+) or without (−) purified bovine brain tubulin and fractionated by native gel electrophoresis to assay for the presence of complexes formed between the labeled β-tubulin and folding cofactors present in the reticulocyte lysate. Results from reactions containing wild-type β-tubulin cDNA (lanes 1 and 2) and mutant cDNAs encoding a 12 amino acid deletion characteristic of strain 6H2 (lanes 3 and 4), an L187R mutation from strain A5 (lanes 5 and 6), and a Y398C mutation from strain 5L1 (lanes 7 and 8) are shown. Complexes formed between β-tubulin and CCT (CCTβ), cofactor D (Fdβ), cofactor A (Faβ), and α-tubulin (αβ) are indicated. Panel B, Reconstituted in vitro folding assay. [35S]methionine labeled wild-type (WT) and mutant Y398C HAβ1-tubulin were produced in E. coli, purified, denatured, and diluted into an in vitro assay containing some or all of the components required to reconstitute the tubulin heterodimer assembly pathway (CCT, ATP, GTP, folding cofactors, and native bovine brain tubulin). The reaction products were resolved by native gel electrophoresis and detected by autoradiography. Complexes formed when HAβ1-tubulin was incubated with CCT alone (lanes 1 and 4), CCT + Fd (lanes 2 and 5), or CCT + Fd + FC + Fe + purified tubulin (i.e., the complete cocktail) (lanes 3 and 6). Panel C, CCT cycles the HAβ1-tubulin. The reactions contained purified, denatured HAβ1-tubulin plus CCT alone (lanes 1 and 4), CCT + a 10X excess of Hsp60 + ATP (lanes 2 and 5), or CCT + Hsp60 + ATP + glucose + hexokinase (lanes 3 and 6). The complex between HAβ1-tubulin and Hsp60 (Hsp60β) is labeled to the left of panel B.
The presence of a band in assembly defective tubulin transcription/translation reactions with the approximate mobility of the αβ heterodimer was surprising given our inability to detect heterodimer formation in vivo by immunoprecipitation (Fig. 4). To determine whether this was indeed heterodimer, purified bovine brain tubulin was added to each of the in vitro translated products and the incubation was continued for 30 minutes. In the case of the wild-type protein, HAβ1-tubulin was efficiently exchanged into the excess unlabeled heterodimers (Fig. 6A, lane 2). However, this did not occur in the case of the assembly defective HAβ1-tubulins (Fig. 6A, lanes 4, 6, and 8). Nonetheless, when each of the reactions was analyzed by SDS-PAGE, comparable amounts of in vitro synthesized tubulin were found to be present in each of the lanes, arguing against the possibility that differences seen were due to poor translation or degradation (data not shown). We conclude that the assembly defective HAβ1-tubulins are incapable of forming heterodimers both in vivo (Fig. 4) and in vitro (Fig. 6A).
To establish whether or not any of the weak bands common to the three assembly defective HAβ1-tubulins analyzed in Fig. 6A represent complexes with the cofactors required for de novo tubulin heterodimer assembly, we tested one of the mutant tubulins using in vitro reconstituted folding reactions containing purified CCT, ATP, GTP and one or more of the components that are essential for the de novo generation of heterodimers. A control reaction using wild-type HAβ1 resulted in the expected interaction with CCT when the chaperonin was present alone (Fig. 6B, lane 1). Inclusion of cofactor D resulted in the generation of a cofactor D/β-tubulin complex (Fig. 6B, lane 2), while the presence of CCT, cofactors C, D, E, and unlabeled native tubulin, (all of which are essential for de novo heterodimer assembly) produced labeled native α/β-tubulin heterodimers (Fig. 6B, lane 3). In contrast, HAβ1-tubulin containing a Y398C substitution still formed a complex with CCT (Fig. 6B, lane 4), but this mutant was unable to form the cofactor D/HAβ1-tubulin complex (Fig. 6B, lane 5) or become incorporated into native heterodimer (Fig. 6B, lane 6).
The ability of assembly defective HAβ1 to form a complex with CCT while failing to yield a complex with cofactor D raised the question as to whether it was capable of undergoing normal cycles of ATP-dependent binding and release from the chaperonin. To test this possibility, unfolded HAβ1-tubulin was presented to CCT for 10 min at 4 °C in the absence of nucleotide in order to allow complex formation (Fig. 6C, lanes 1 and 4). ATP and a 10-fold molar excess of Hsp60 (with respect to CCT) was then added as a trap for the capture of cycling intermediates, and the reactions were warmed to 30 °C for 30 min. Under these conditions, the trap captured wild-type (Fig. 6C, lane 2) and assembly defective (Fig. 6C, lane 5) HAβ1-tubulin with indistinguishable efficiency. As a control, a parallel reaction with Hsp60 was carried out in the presence of glucose and hexokinase to quench ATP. In this case, no capture of wild-type or assembly defective HAβ1-tubulin could be seen (Fig. 6C, lanes 3 and 6). We conclude that assembly defective HAβ1-tubulin is efficiently cycled by CCT, but the intermediates produced are not competent for capture by cofactor D, and hence cannot participate in productive heterodimer formation. Consistent with this conclusion, we were able to immunoprecipitate cofactor D complexes with wild-type but not assembly defective β-tubulin using extracts of cells transfected so as to overexpress a GFP-cofactor D chimeric protein (Supplemental Figure 1).
Proteasomal Degradation of Assembly Defective Tubulin
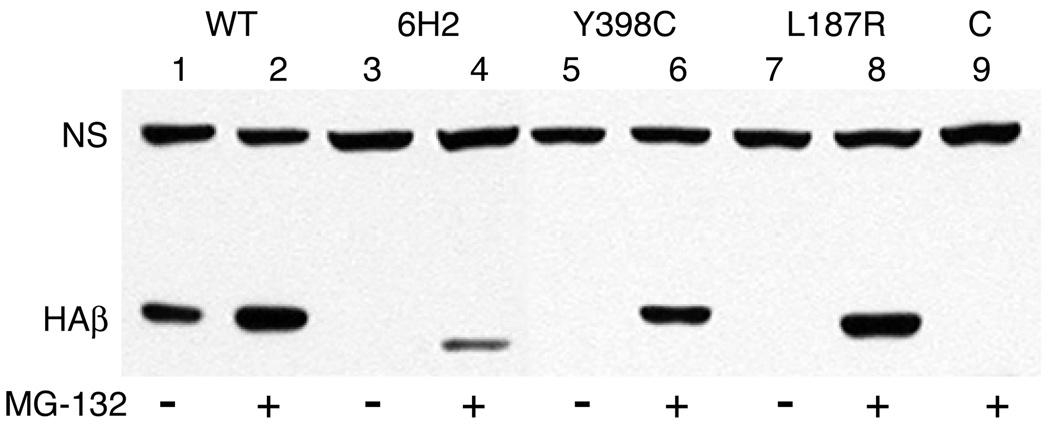
Little is currently known about how cells recognize defective tubulins and target them for degradation. To test whether the assembly defective tubulins in our revertant strains are degraded by the proteasome, we transfected wild-type CHO cells with HAβ1-tubulin cDNAs containing the 12 amino acid deletion and each of the two point mutations. Stably transfected cell lines were selected, treated for 8 h with or without the proteasomal inhibitor MG132, and analyzed by western blots using an antibody that recognizes the HA tag (Fig. 7). We found that the abundance of each of the assembly defective HAβ1-tubulins was dramatically increased in the presence of MG132, indicating that mutant tubulins are degraded by the proteasome. Interestingly, the abundance of transfected wild-type HAβ1-tubulin was also elevated, indicating that overexpressed wild-type subunits are also degraded by the proteasome.
Fig. 7.
Inhibition of HAβ1-tubulin degradation. CHO cells were stably transfected with wild-type HAβ1-tubulin cDNA (WT, lanes 1 and 2) or with the same cDNA encoding a 12 amino acid deletion (6H2, lanes 3 and 4), a Y398C mutation (lanes 5 and 6), or an L187R mutation (lanes 7 and 8). The transfected cell lines were then treated with (+) or without (−) the proteasomal inhibitor MG132 for 8 h and lysed in SDS. Proteins were analyzed by Western blotting with a mouse antibody to the HA tag. Bands representing transfected HAβ1-tubulin (HAβ) and a nonspecific endogenous CHO protein that cross reacts with the antibody (NS) and acts as a loading control are labeled. “C” (lane 9) represents a control lysate from non-transfected cells that were treated with MG132.
DISCUSSION
The isolation and characterization of revertants of drug resistant CHO cells is providing new approaches to study microtubule biology. Our prior characterization of colcemid sensitive revertants of Cmd 4, a colcemid resistant CHO cell line with hyperstable microtubules, demonstrated that loss of mutant tubulin expression due to deletions and frameshift mutations represents the most common mechanism of reversion (19). In addition, cells can revert by the acquisition of intra-allelic point mutations that destabilize microtubules, or that increase colcemid binding (18). Here, we describe a newly discovered mechanism of reversion, namely, the acquisition of mutations that interfere with productive tubulin folding and thereby prevent its assembly into heterodimers and hasten its degradation. This is a relatively common mechanism that applies to approximately 20% of revertant cell lines. Thus, selecting revertants of drug resistant cells provides an efficient method for the isolation of mutations that affect tubulin folding. Mutations that confer this phenotype can include single amino acid substitutions, in-frame deletions, and truncations.
Although the pathway for the folding of newly synthesized wild-type tubulin subunits has been worked out in some detail, little is known about the mechanisms by which cells detect and degrade defective tubulin proteins. The studies described here suggest that recognition of misfolded tubulin occurs at an intermediate stage in the folding pathway. Assembly defective β-tubulin polypeptides bind to CCT with a stoichiometry that reflects their synthesis rates relative to wild-type subunits, but they do not appear in other discernible complexes. In vitro pulse/chase experiments further indicate that newly synthesized assembly defective proteins become associated with and are discharged from CCT with the same kinetics as wild-type protein (data not shown). Moreover, in vitro reconstitution with one of the mutations confirms that assembly defective tubulin cycles through the CCT complex in the same manner as wild-type (Fig. 6C). It thus appears that the assembly defective forms of β-tubulin proceed through the early steps of the folding pathway, but they are unable to interact with the cofactors that are required for their assembly into heterodimers.
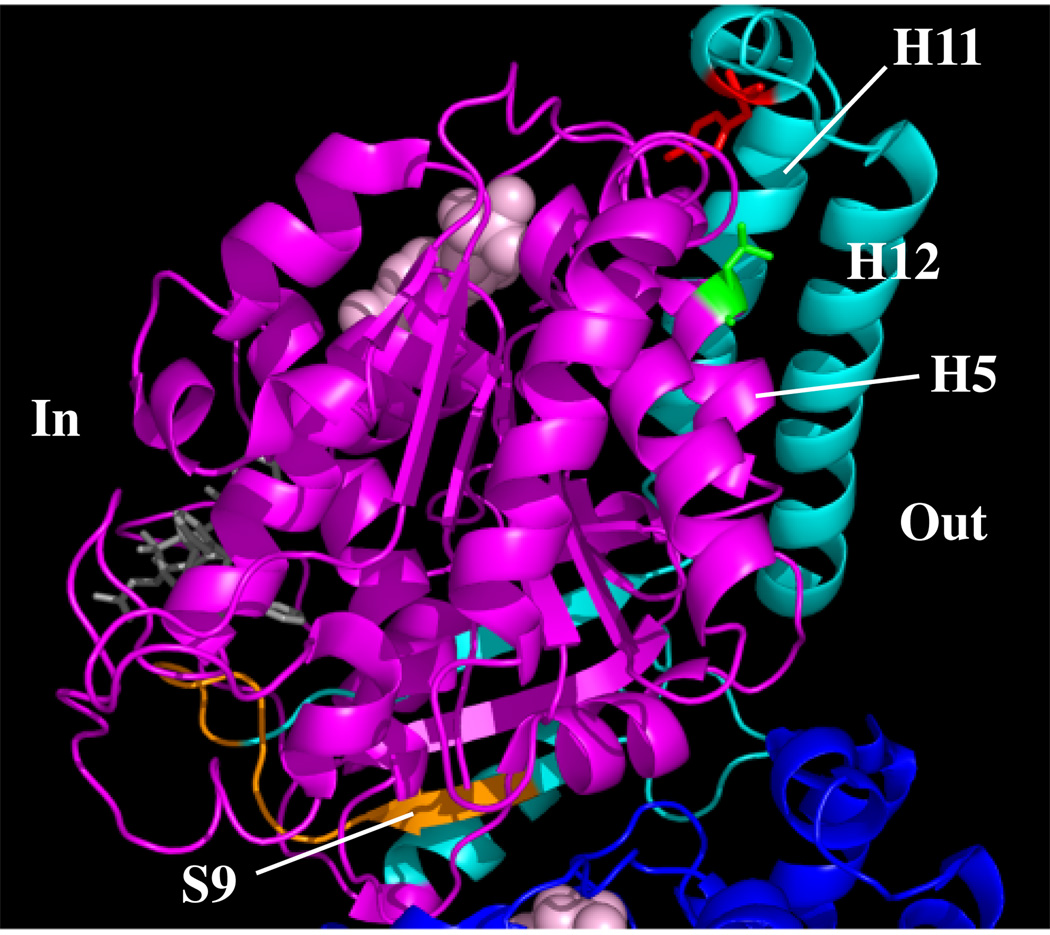
We did not note any differences in behavior among two different point mutations, a small inframe deletion, and a C-terminal truncation, suggesting that both subtle and gross changes in structure may be recognized by the same quality control mechanism. Revertant 6H3 has a large deletion (Fig. 8, cyan color) that eliminates helices H11 and H12 that form a “backbone” on the outer surface of microtubule protofilaments, helix H10 located near the intradimer interface, sheets S9 and S10, and all of the intervening loops. It is no surprise that such massive changes would result in misfolding. The smaller 12 amino acid deletion in revertant 6H2 (Fig. 8, orange color) also affects S9 near the intradimer interface as well as the following loop. The loss of these 12 residues again appears to cause sufficient structural changes to preclude normal folding. Of greatest interest are the two point mutations that are approximately 6 Å apart in the tertiary structure. The 5L1 mutation produces a Y398C substitution (Fig. 8, red color) in the small helix H11’ that interrupts a loop connecting the large helices H11 and H12. The L187R substitution (Fig. 8, green color) in revertant A5 is located in helix H5 that is also very close to helices H11 and H12. It is conceivable that each of these 2 mutations could affect the conformation or position of H11 and H12 and thereby interfere with the binding of a critical cofactor in the folding pathway. It has been found, for example, that cofactor A has a highly extended helical conformation (33), and we speculate that it could potentially interact with H11 and H12, protecting the quasi-native protein from the degradation pathway. Evidence to support this idea has recently come from the observation that knockdown of cofactor A using siRNA causes a large reduction in cellular tubulin (34). The inability of either cofactor A or cofactor D to bind properly to the β-tubulin released from CCT might thus trigger its degradation.
Fig. 8.
Location of destabilizing mutations. Mutations that interfere with β-tubulin folding are shown on the molecular structure reported by Lowe et al. (37). The β subunit is shown in magenta and a small portion of the α subunit is shown in dark blue. Regions affected by the 6H3 truncation are in cyan, the 6H2 12 amino acid deletion is in orange, the L187R substitution is in green, and the Y398C substitution is in red. The tubulin is oriented with the region that faces the interior of the microtubule to the left (In) and the region that faces the exterior to the right (Out). Helices H5, H11, and H12; and β-sheet S9 are also labeled. The structure was drawn with MacPyMol (www.pymol.org).
The enhanced stability of assembly defective β-tubulin in the presence of MG132 indicates that mutant tubulin degradation proceeds through the proteasome. Currently, little else is known about the degradative pathway for aberrant forms of tubulin. Feng and associates have reported that parkin, a ubiquitin E-3 ligase implicated in Parkinson’s disease, enhances the ubiquitination and degradation of tubulin (35). Because those studies were carried out with cells containing wild-type tubulin, the authors concluded that a small percentage of wild-type subunits are misfolded following synthesis, a conclusion that is supported by our observation that not only mutant, but also overexpressed wild-type tubulin, is stabilized in cells treated with MG132, albeit to a lesser extent. It thus appears that the surveillance system that recognizes and degrades mutant forms of β-tubulin may also target misfolded wild-type subunits. This surveillance system appears to be distinct from, and acts at an earlier step than the quality control mechanism that monitors the ability of tubulin heterodimers to hydrolyse GTP (12).
Tubulin is essential and therefore some minimum level of the protein must be required for cell survival. At present, however, it is unclear whether cellular tubulin is maintained at a critical concentration or whether it is present in excess of the minimum needed for normal growth. Our studies demonstrate that loss of half of the β1-tubulin in the revertant cell lines produces the expected 35% decrease in total cellular tubulin, yet does not interfere with cell proliferation. CHO cells in culture must therefore have more tubulin than they need for survival, and significant reductions in tubulin do not trigger mechanisms to maintain tubulin at normal wild-type levels. Our data further indicate that CHO cells do not require microtubule abundance to be maintained at a constant level: revertant cells have 35% less microtubule polymer and 35% less free tubulin than wild-type cells. These observations support the idea that CHO cells can tolerate considerable fluctuations in tubulin and microtubule abundance with little or no adaptive response or ill effects, reinforcing a similar conclusion based on other mutant analyses (36). The lack of compensatory mechanisms to deal with reductions in tubulin content suggests that therapies targeted toward reducing cellular tubulin might prove to be effective treatments for cancer.
Supplementary Material
Footnotes
The authors thank Don Cleveland, Jeanette Bulinski, and William Margolin for generously providing us with antibodies needed to carry out these studies. This work was supported by grant CA85935 (F. C.) and grant DK47234 (N. J. C.) from the National Institutes of Health.
The abbreviations used are: CCT, cytosolic chaperonin containing TCP-1; CHO, Chinese hamster ovary; HA, hemagglutinin; MTB, microtubule buffer; PAGE, polyacrylamide gel electrophoresis; tet, tetracycline.
Contributor Information
Yaqing Wang, Department of Integrative Biology and Pharmacology, The University of Texas Medical School, Houston, TX 77030.
Guoling Tian, Department of Biochemistry, New York University Medical Center, New York, NY 10016.
Nicholas J. Cowan, Department of Biochemistry, New York University Medical Center, New York, NY 10016
Fernando Cabral, Department of Integrative Biology and Pharmacology, The University of Texas Medical School, Houston, TX 77030.
REFERENCES
- 1.Hansen WJ, Cowan NJ, Welch WJ. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 3.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Embo J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 5.Spiess C, Meyer AS, Reissmann S, Frydman J. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 7.Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhamidipati A, Lewis SA, Cowan NJ. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowan NJ, Lewis SA. Adv Protein Chem. 2001;59:73–104. doi: 10.1016/s0065-3233(01)59003-8. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan KF. Ann. Rev. Cell Biol. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- 11.Luduena RF. Internatl. Rev. Cytol. 1998;178:207–275. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- 12.Tian G, Bhamidipati A, Cowan NJ, Lewis SA. J Biol Chem. 1999;274:24054–24058. doi: 10.1074/jbc.274.34.24054. [DOI] [PubMed] [Google Scholar]
- 13.Cabral F. Drug Resistance Updates. 2000;3:1–6. doi: 10.1054/drup.2000.0172. [DOI] [PubMed] [Google Scholar]
- 14.Orr GA, Verdier-Pinard P, McDavid H, Horwitz SB. Oncogene. 2003;22:7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabral F, Abraham I, Gottesman MM. Mol. Cell. Biol. 1982;2:720–729. doi: 10.1128/mcb.2.6.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabral F, Sobel ME, Gottesman MM. Cell. 1980;20:29–36. doi: 10.1016/0092-8674(80)90231-7. [DOI] [PubMed] [Google Scholar]
- 17.Hari M, Wang Y, Veeraraghavan S, Cabral F. Mol. Cancer Ther. 2003;2:597–605. [PubMed] [Google Scholar]
- 18.Wang Y, Veeraraghavan S, Cabral F. Biochemistry. 2004;43:8965–8973. doi: 10.1021/bi049637b. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Cabral F. Biochim. Biophys. Acta. 2005;1744:245–255. doi: 10.1016/j.bbamcr.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Boggs B, Cabral F. Mol. Cell. Biol. 1987;7:2700–2707. doi: 10.1128/mcb.7.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli UK. Nature (Lond.) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Cabral F, Schatz G. Methods Enzymol. 1979;56:602–613. doi: 10.1016/0076-6879(79)56057-1. [DOI] [PubMed] [Google Scholar]
- 23.Towbin H, Staehelin T, Gordon J. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minotti AM, Barlow SB, Cabral F. J. Biol. Chem. 1991;266:3987–3994. [PubMed] [Google Scholar]
- 25.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 26.Lopata MA, Cleveland DW. J. Cell Biol. 1987;105:1707–1720. doi: 10.1083/jcb.105.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawada T, Cabral F. J. Biol. Chem. 1989;264:3013–3020. [PubMed] [Google Scholar]
- 28.Ahmad S, Singh B, Gupta RS. Biochim. Biophys. Acta. 1991;1090:252–254. doi: 10.1016/0167-4781(91)90112-y. [DOI] [PubMed] [Google Scholar]
- 29.Spiegelman BM, Penningroth SM, Kirschner MW. Cell. 1977;12:587–600. doi: 10.1016/0092-8674(77)90259-8. [DOI] [PubMed] [Google Scholar]
- 30.Boggs BA, Minotti AM, Loeb LM, Cook R, Cabral F. J. Biol. Chem. 1988;263:14566–14573. [PubMed] [Google Scholar]
- 31.Chen X, Sullivan DS, Huffaker TC. Proc. Natl. Acad. Sci. USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ursic D, Sedbrook JC, Himmel KL, Culbertson MR. Mol. Biol. Cell. 1994;5:1065–1080. doi: 10.1091/mbc.5.10.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guash A, Aloria K, Perez R, Avila J, Zabala JC, Coll M. J. Mol. Biol. 2002;318:1139–1149. doi: 10.1016/S0022-2836(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 34.Nolasco S, Bellido J, Gancalves J, Zabala JC, Soares H. FEBS Lett. 2005;579:3515–3524. doi: 10.1016/j.febslet.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Ren Y, Zhao J, Feng J. J. Neurosci. 2003;23:3316–3324. doi: 10.1523/JNEUROSCI.23-08-03316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barlow SB, Gonzalez-Garay ML, Cabral F. J. Cell Sci. 2002;115:3469–3478. doi: 10.1242/jcs.115.17.3469. [DOI] [PubMed] [Google Scholar]
- 37.Lowe J, Li H, Downing KH, Nogales E. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.