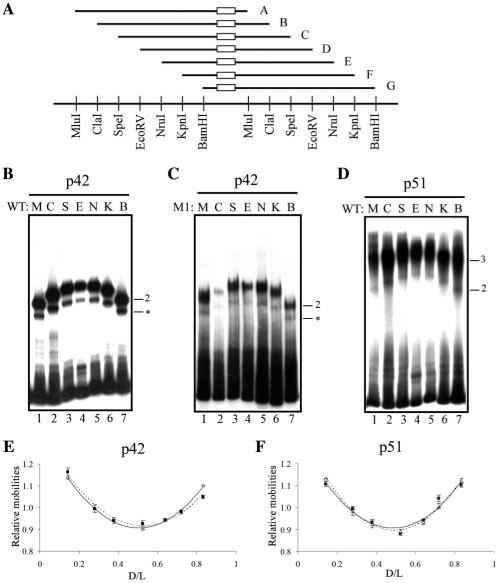
Figure 4.
Determination of DNA bending amplitude induced by p51 and p42 Ets-1 isoforms. (A) Schematic representation of DNA probes used for the circular permutation assays. WT or M1 are symbolized by an open rectangle in probes A-G that correspond to M, C, S, E, N, K and B nomenclature according to the restriction sites. Restriction enzyme sites are given below the probe representations. (B, C, D) Gel shift assays performed with p42 on WT probe (B), p42 on M1 probe (C) and p51 on WT probe (D), as described in A. Binary (2) and ternary (3) complexes are indicated. Non-relevant complexes are indicated by an asterisk. (E, F) Relative mobility as a function of D/L; D is the number of base pairs separating the 5′ end of the probe and the EBS centers (center of M1 or center of WT palindrome); L is the length of the probe, expressed in base pairs. The lines correspond to the best second-order polynomial fit. In E, open diamond: p42-WT and filled square: p42-M1. In F, unfilled triangle: p51–WT and filled square: p51–WT–p51.