Figure 5.
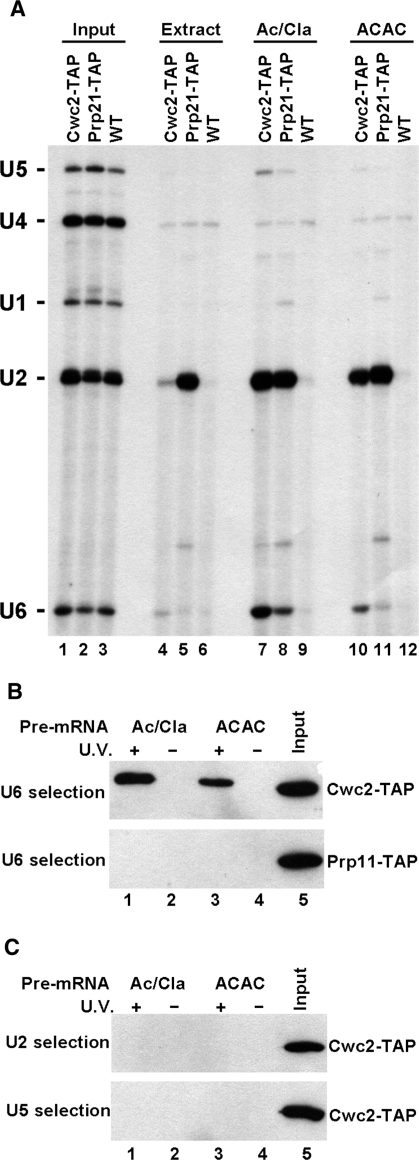
Cwc2 is only snRNA-associated within spliceosomes and contacts the U6 snRNA directly in first and second step spliceosomes. (A) Yeast whole-cell extract was made from yeast strains containing TAP-tagged Cwc2 (Cwc2-TAP), TAP-tagged Prp21 (Prp21-TAP) or a non-TAP-tagged wild-type (WT) strain. Total RNA was isolated from 1.25% of extract used in the precipitations from extracts and splicing reactions, then subjected to primer extension with primers complementary to the U1, U2, U4, U5 and U6 snRNAs (lanes 1–3, Input). Extract alone (lanes 4–6, Extract), extract incubated under splicing conditions with an ACT1 pre-mRNA truncated after the branchpoint sequence (lanes 7–9, Ac/Cla) or extract incubated under splicing conditions with an ACT1 pre-mRNA with a 3′-splice site mutation (lanes 10–12, ACAC) were precipitated with IgG-coated beads. RNA extracted from the beads was subjected to primer extension with primers complementary to the U1, U2, U4, U5 and U6 snRNAs. The positions of each snRNA are shown on the left. (B) Yeast whole-cell extract was made from yeast strains containing TAP-tagged Cwc2 (Cwc2-TAP) (top panel) and TAP-tagged Prp11 (Prp11-TAP) (bottom panel). Splicing reactions with Ac/Cla (lanes 1 and 2) or ACAC (lanes 3 and 4) pre-mRNA were UV irradiated (lanes 1 and 3) before selection with a biotinylated oligonucleotide complementary to U6 snRNA, under denaturing conditions. Cwc2-TAP or Prp11-TAP were detected with the anti-TAP antibody by immunoblotting. As a control, identical splicing reactions were subjected to selection of U6 under the same denaturing conditions without previous UV irradiation (lanes 2 and 4). Lane 5 shows 1% of the TAP-tagged extract used in splicing reactions. (C) As in (B) except selection from Cwc2-TAP extract splicing reactions after crosslinking utilized biotinylated oligonucleotides complementary to U2 snRNA (top panel) or U5 snRNA (bottom panel).