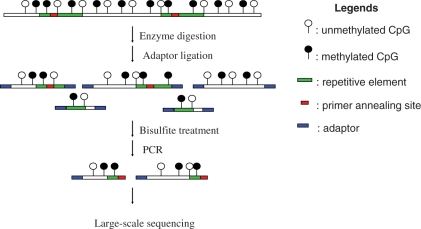
Figure 1.
Diagram of experimental design. Genomic DNA is first digested with a methylation insensitive enzyme, ligated to adaptors and then subjected to bisulfite treatment. Bisulfite treated DNA is amplified with adaptor and primer specific for the targeted repeat elements. PCR products contain repeat sequence and flanking unique genomic sequence. Lastly, the repeat-containing amplicon library thus constructed is subjected to large-scale sequencing. The methylated cytosines are indicated with the filled circles while the unmethylated cytosines are indicated with the open circles. The green and red segments represent the repeat elements with the red segment indicating the region to which the primer anneals. The adaptors are indicated by blue boxes.