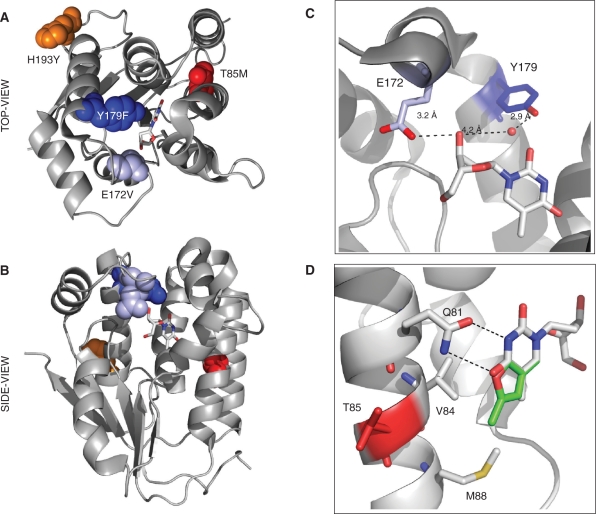
Figure 3.
Structure model of R4.V3. The locations of the four mutations are highlighted in DmdNK with thymidine, bound in the phosphoryl acceptor-binding site (PDB access no.: 1OT3 (40)). The top (A) and side view (B) of DmdNK show the close proximity of E172V (light blue) and Y179F (dark blue) to the substrate while H193Y (orange) and T85M (red) are located in more distant locations to the active site. (C) The close-up view of the active site shows the direct hydrogen-bonding interactions of E172 with the 3′-OH group of the substrate's ribose moiety. The OH group of Y179 contacts the same position on the substrate via bridging water. (D) The modeled overlay of thymidine (gray) and fT (green) in the active site of DmdNK indicates a potential steric clash of the fluorescent substrate's furano moiety with V84 and M88. The T85M mutation is believed to cause a slight conformational shift of the helical region, creating a more favorable binding pocket for substrates with modifications in the C4/C5 position of the nucleobase. Figures were created with PyMOL software (DeLano Sci., Palo Alto, CA).