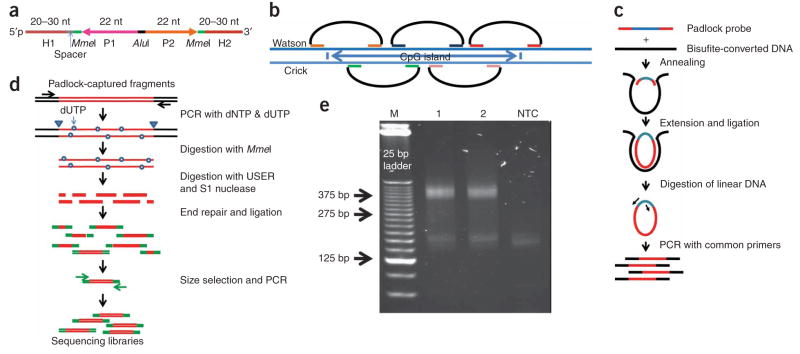
Figure 1.
Targeted bisulfite sequencing with padlock probes. (a) Each padlock probe has a common linker sequence flanked by two target-specific capturing arms (H1 and H2). H1 and H2 are melting temperature–normalized, and a spacer sequence is included to normalize probe lengths. The linker sequence contains priming sites (AP1 and AP2) for universal primers, two MmeI sites and a central AluI recognition site. (b) A CpG island (or other target regions) is covered by multiple padlock probes targeting partially overlapped regions on alternating strands. (c) A library of padlock probes is annealed to bisulfite-converted genomic DNA (black) and the 3′ ends are extended and ligated with the 5′ end. After removal of linear DNAs with exonucleases, all circularized padlock probes are PCR-amplified using a pair of common primers. (d) To generate a shotgun sequencing library, amplicons were reamplified in the presence of dUTP, digested with MmeI, and then with USER and S1 nuclease. Digested amplicons are end repaired and ligated with Solexa sequencing adaptors (green). Ligated products are then selected by size and amplified by PCR to generate the shotgun sequencing library. (e) Gel electrophoresis analysis of the padlock-captured products from two independent capturing reactions (1 and 2) and a no-template control. Expected amplicon size is in the range of 344–394 bp, which includes capturing targets (175–225 bp), capturing arms (58 bp) and amplification primers (111 bp). NTC, no-template control.